[English] 日本語
 Yorodumi
Yorodumi- SASDCY6: T2 in b-DDM micelle (SEC-SAXS) (Mechanosensitive channel T2, T2 +... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: SASBDB / ID: SASDCY6 |
|---|---|
 Sample Sample | T2 in b-DDM micelle (SEC-SAXS)
|
| Function / homology | Mechanosensitive ion channel MscS, archaea/bacteria type / Mechanosensitive ion channel MscS, transmembrane-2 / Mechanosensitive ion channel MscS / Mechanosensitive ion channel, beta-domain / mechanosensitive monoatomic ion channel activity / LSM domain superfamily / membrane / Mechanosensitive ion channel MscS domain-containing protein Function and homology information Function and homology information |
| Biological species |   Thermoplasma volcanium (strain ATCC 51530 / DSM 4299 / JCM 9571 / NBRC 15438 / GSS1) (archaea) Thermoplasma volcanium (strain ATCC 51530 / DSM 4299 / JCM 9571 / NBRC 15438 / GSS1) (archaea)Synthetic compound (others) |
 Citation Citation |  Date: 2018 Feb Date: 2018 FebTitle: Saposin Lipid Nanoparticles: A Highly Versatile and Modular Tool for Membrane Protein Research Authors: Flayhan A / Mertens H / Ural-Blimke Y / Martinez Molledo M / Svergun D |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
-Data source
| SASBDB page |  SASDCY6 SASDCY6 |
|---|
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- External links
External links
| Related items in Molecule of the Month |
|---|
-Models
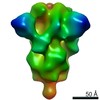
| Model #1498 |  Type: dummy / Software: (2.9.0) / Radius of dummy atoms: 3.50 A / Symmetry: P7 / Chi-square value: 3.360  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
|---|
- Sample
Sample
 Sample Sample | Name: T2 in b-DDM micelle (SEC-SAXS) / Entity id: 786 / 789 |
|---|---|
| Buffer | Name: 20 mM Tris 150 mM NaCl 5 % glycerol 0.03 % DDM / pH: 7.5 |
| Entity #786 | Name: T2 / Type: protein / Description: Mechanosensitive channel T2 / Formula weight: 32.887 / Num. of mol.: 7 Source: Thermoplasma volcanium (strain ATCC 51530 / DSM 4299 / JCM 9571 / NBRC 15438 / GSS1) References: UniProt: Q97AH5 Sequence: MDRAKGKSIA VYVGSMIVLI IVLLALLYVL SLFHIVPIKF SRILYAVVIG LVIYAIVRIV TKYVERFISL HSEFKHLKYI VFIISLLGYF IISLAVLAAL GIDVSSVILG SAFLSAIIGL AAQSVLANVF GGLFLSIVRP FKLGDHIVVN TWQFPVSFPS YPPKYFSRDF ...Sequence: MDRAKGKSIA VYVGSMIVLI IVLLALLYVL SLFHIVPIKF SRILYAVVIG LVIYAIVRIV TKYVERFISL HSEFKHLKYI VFIISLLGYF IISLAVLAAL GIDVSSVILG SAFLSAIIGL AAQSVLANVF GGLFLSIVRP FKLGDHIVVN TWQFPVSFPS YPPKYFSRDF IEGSIYRGIV SDISINYTSI ALESGDVVKI PNNIMIQAAI TLRHGSVTVQ ARYEVPKYIN FNDIYNKIVD EVGTLPGARN IGVMIDETTV NTYIILVRGD FQGEDADLVR SQILQKVIGI VEPLKKQ |
| Entity #789 | Name: b-DDM / Type: other / Description: n-Dodecyl-β-D-Maltopyranoside / Formula weight: 0.511 / Source: Synthetic compound |
-Experimental information
| Beam | Instrument name: PETRA III P12 / City: Hamburg / 国: Germany  / Type of source: X-ray synchrotron / Wavelength: 0.124 Å / Dist. spec. to detc.: 3.1 mm / Type of source: X-ray synchrotron / Wavelength: 0.124 Å / Dist. spec. to detc.: 3.1 mm | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 2M | ||||||||||||||||||||||||||||||||||||||||||
| Scan |
| ||||||||||||||||||||||||||||||||||||||||||
| Distance distribution function P(R) |
| ||||||||||||||||||||||||||||||||||||||||||
| Result |
|
 Movie
Movie Controller
Controller