+ Open data
Open data
- Basic information
Basic information
| Entry | Database: SASBDB / ID: SASDAH8 |
|---|---|
 Sample Sample | Human Filamin A Ig-like domains 20-21* truncation (2141-2329)
|
| Function / homology |  Function and homology information Function and homology informationregulation of membrane repolarization during atrial cardiac muscle cell action potential / regulation of membrane repolarization during cardiac muscle cell action potential / establishment of Sertoli cell barrier / Myb complex / formation of radial glial scaffolds /  glycoprotein Ib-IX-V complex / adenylate cyclase-inhibiting dopamine receptor signaling pathway / positive regulation of integrin-mediated signaling pathway / cytoplasmic sequestering of protein / tubulin deacetylation ...regulation of membrane repolarization during atrial cardiac muscle cell action potential / regulation of membrane repolarization during cardiac muscle cell action potential / establishment of Sertoli cell barrier / Myb complex / formation of radial glial scaffolds / glycoprotein Ib-IX-V complex / adenylate cyclase-inhibiting dopamine receptor signaling pathway / positive regulation of integrin-mediated signaling pathway / cytoplasmic sequestering of protein / tubulin deacetylation ...regulation of membrane repolarization during atrial cardiac muscle cell action potential / regulation of membrane repolarization during cardiac muscle cell action potential / establishment of Sertoli cell barrier / Myb complex / formation of radial glial scaffolds /  glycoprotein Ib-IX-V complex / adenylate cyclase-inhibiting dopamine receptor signaling pathway / positive regulation of integrin-mediated signaling pathway / cytoplasmic sequestering of protein / tubulin deacetylation / actin crosslink formation / glycoprotein Ib-IX-V complex / adenylate cyclase-inhibiting dopamine receptor signaling pathway / positive regulation of integrin-mediated signaling pathway / cytoplasmic sequestering of protein / tubulin deacetylation / actin crosslink formation /  blood coagulation, intrinsic pathway / protein localization to bicellular tight junction / OAS antiviral response / positive regulation of actin filament bundle assembly / positive regulation of neuron migration / positive regulation of potassium ion transmembrane transport / Cell-extracellular matrix interactions / early endosome to late endosome transport / blood coagulation, intrinsic pathway / protein localization to bicellular tight junction / OAS antiviral response / positive regulation of actin filament bundle assembly / positive regulation of neuron migration / positive regulation of potassium ion transmembrane transport / Cell-extracellular matrix interactions / early endosome to late endosome transport /  apical dendrite / positive regulation of neural precursor cell proliferation / cell-cell junction organization / positive regulation of platelet activation / protein localization to cell surface / Fc-gamma receptor I complex binding / negative regulation of transcription by RNA polymerase I / apical dendrite / positive regulation of neural precursor cell proliferation / cell-cell junction organization / positive regulation of platelet activation / protein localization to cell surface / Fc-gamma receptor I complex binding / negative regulation of transcription by RNA polymerase I /  wound healing, spreading of cells / megakaryocyte development / GP1b-IX-V activation signalling / cortical cytoskeleton / positive regulation of axon regeneration / receptor clustering / SMAD binding / RHO GTPases activate PAKs / actin filament bundle / wound healing, spreading of cells / megakaryocyte development / GP1b-IX-V activation signalling / cortical cytoskeleton / positive regulation of axon regeneration / receptor clustering / SMAD binding / RHO GTPases activate PAKs / actin filament bundle /  brush border / semaphorin-plexin signaling pathway / brush border / semaphorin-plexin signaling pathway /  cilium assembly / cilium assembly /  mitotic spindle assembly / potassium channel regulator activity / mitotic spindle assembly / potassium channel regulator activity /  epithelial to mesenchymal transition / blood vessel remodeling / axonal growth cone / heart morphogenesis / positive regulation of substrate adhesion-dependent cell spreading / epithelial to mesenchymal transition / blood vessel remodeling / axonal growth cone / heart morphogenesis / positive regulation of substrate adhesion-dependent cell spreading /  regulation of cell migration / release of sequestered calcium ion into cytosol / dendritic shaft / regulation of cell migration / release of sequestered calcium ion into cytosol / dendritic shaft /  protein kinase C binding / protein localization to plasma membrane / G protein-coupled receptor binding / protein kinase C binding / protein localization to plasma membrane / G protein-coupled receptor binding /  actin filament / synapse organization / mRNA transcription by RNA polymerase II / actin filament / synapse organization / mRNA transcription by RNA polymerase II /  trans-Golgi network / establishment of protein localization / negative regulation of DNA-binding transcription factor activity / negative regulation of protein catabolic process / cerebral cortex development / Z disc / trans-Golgi network / establishment of protein localization / negative regulation of DNA-binding transcription factor activity / negative regulation of protein catabolic process / cerebral cortex development / Z disc /  small GTPase binding / small GTPase binding /  platelet aggregation / platelet aggregation /  kinase binding / positive regulation of protein import into nucleus / kinase binding / positive regulation of protein import into nucleus /  actin filament binding / cell-cell junction / actin filament binding / cell-cell junction /  actin cytoskeleton / Platelet degranulation / negative regulation of neuron projection development / actin cytoskeleton / Platelet degranulation / negative regulation of neuron projection development /  GTPase binding / GTPase binding /  perikaryon / actin cytoskeleton organization / postsynapse / perikaryon / actin cytoskeleton organization / postsynapse /  angiogenesis / positive regulation of canonical NF-kappaB signal transduction / DNA-binding transcription factor binding / transmembrane transporter binding / protein stabilization / angiogenesis / positive regulation of canonical NF-kappaB signal transduction / DNA-binding transcription factor binding / transmembrane transporter binding / protein stabilization /  cadherin binding / cadherin binding /  focal adhesion / glutamatergic synapse / focal adhesion / glutamatergic synapse /  nucleolus / negative regulation of apoptotic process / perinuclear region of cytoplasm / protein homodimerization activity / nucleolus / negative regulation of apoptotic process / perinuclear region of cytoplasm / protein homodimerization activity /  RNA binding / extracellular exosome / extracellular region / RNA binding / extracellular exosome / extracellular region /  membrane / membrane /  nucleus / nucleus /  plasma membrane / plasma membrane /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function |
| Biological species |   Homo sapiens (human) Homo sapiens (human) |
 Citation Citation |  Journal: PLoS One / Year: 2015 Journal: PLoS One / Year: 2015Title: Flexible Structure of Peptide-Bound Filamin A Mechanosensor Domain Pair 20-21. Authors: Jonne Seppälä / Helena Tossavainen / Nebojsa Rodic / Perttu Permi / Ulla Pentikäinen / Jari Ylänne /  Abstract: Filamins (FLNs) are large, multidomain actin cross-linking proteins with diverse functions. Besides regulating the actin cytoskeleton, they serve as important links between the extracellular matrix ...Filamins (FLNs) are large, multidomain actin cross-linking proteins with diverse functions. Besides regulating the actin cytoskeleton, they serve as important links between the extracellular matrix and the cytoskeleton by binding cell surface receptors, functioning as scaffolds for signaling proteins, and binding several other cytoskeletal proteins that regulate cell adhesion dynamics. Structurally, FLNs are formed of an amino terminal actin-binding domain followed by 24 immunoglobulin-like domains (IgFLNs). Recent studies have demonstrated that myosin-mediated contractile forces can reveal hidden protein binding sites in the domain pairs IgFLNa18-19 and 20-21, enabling FLNs to transduce mechanical signals in cells. The atomic structures of these mechanosensor domain pairs in the resting state are known, as well as the structures of individual IgFLN21 with ligand peptides. However, little experimental data is available on how interacting protein binding deforms the domain pair structures. Here, using small-angle x-ray scattering-based modelling, x-ray crystallography, and NMR, we show that the adaptor protein migfilin-derived peptide-bound structure of IgFLNa20-21 is flexible and adopts distinctive conformations depending on the presence or absence of the interacting peptide. The conformational changes reported here may be common for all peptides and may play a role in the mechanosensor function of the site. |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
-Data source
| SASBDB page |  SASDAH8 SASDAH8 |
|---|
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- External links
External links
| Related items in Molecule of the Month |
|---|
-Models
| Model #321 |  Type: dummy / Software: DAMMIF (r4556) / Radius of dummy atoms: 1.40 A / Chi-square value: 0.752 / P-value: 0.249000  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
|---|
- Sample
Sample
 Sample Sample | Name: Human Filamin A Ig-like domains 20-21* truncation (2141-2329) Specimen concentration: 1.00-4.00 |
|---|---|
| Buffer | Name: Tris / Concentration: 20.00 mM / pH: 8 / Composition: 50 mM NaCl, 10mM DTT / Concentration: 20.00 mM / pH: 8 / Composition: 50 mM NaCl, 10mM DTT |
| Entity #190 | Name: FilaminA* / Type: protein / Description: Human Filamin A Ig-like domains 20-21* / Formula weight: 20 / Num. of mol.: 1 / Source: Homo sapiens / References: UniProt: P21333 Sequence: KESITRRRRA PSVANVGSHC DLSLKIPEIS IQDMTAQVTS PSGKTHEAEI VEGENHTYCI RFVPAEMGTH TVSVKYKGQH VPGSPFQFTV GPLGEGGAHK VRAGGPGLER AEAGVPAEFS IWTREAGAGG LAIAVEGPSK AEISFEDRKD GSCGVAYVVQ EPGDYEVSVK FNEEHIPDSP FVVPVASPS |
-Experimental information
| Beam | Instrument name: ESRF BM29 / City: Grenoble / 国: France  / Type of source: X-ray synchrotron / Type of source: X-ray synchrotron Synchrotron / Wavelength: 0.93 Å / Dist. spec. to detc.: 2.43 mm Synchrotron / Wavelength: 0.93 Å / Dist. spec. to detc.: 2.43 mm | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: Pilatus 1M | ||||||||||||||||||||||||||||||
| Scan |
| ||||||||||||||||||||||||||||||
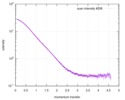
| Distance distribution function P(R) |
| ||||||||||||||||||||||||||||||
| Result | Comments: Filamin A fragment residues 2141-2329 (Ig-like domains).
|
 Movie
Movie Controller
Controller