+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3k2s | ||||||
|---|---|---|---|---|---|---|---|
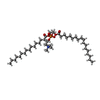
| Title | Solution structure of double super helix model | ||||||
 Components Components | Apolipoprotein A-I Apolipoprotein AI Apolipoprotein AI | ||||||
 Keywords Keywords | LIPID BINDING PROTEIN / super double helix /  amphipathic / amphipathic /  Amyloid / Amyloid /  Amyloidosis / Amyloidosis /  Atherosclerosis / Cholesterol metabolism / Disease mutation / Atherosclerosis / Cholesterol metabolism / Disease mutation /  Glycation / Glycation /  Glycoprotein / HDL / Glycoprotein / HDL /  Lipid metabolism / Lipid transport / Lipid metabolism / Lipid transport /  Lipoprotein / Lipoprotein /  Neuropathy / Neuropathy /  Palmitate / Palmitate /  Secreted / Secreted /  Steroid metabolism / Transport / HIGH DENSITY LIPOPROTEIN Steroid metabolism / Transport / HIGH DENSITY LIPOPROTEIN | ||||||
| Function / homology |  Function and homology information Function and homology informationDefective ABCA1 causes TGD / Scavenging by Class B Receptors / HDL clearance / high-density lipoprotein particle receptor binding / spherical high-density lipoprotein particle / positive regulation of hydrolase activity / negative regulation of response to cytokine stimulus / regulation of intestinal cholesterol absorption / protein oxidation / vitamin transport ...Defective ABCA1 causes TGD / Scavenging by Class B Receptors / HDL clearance / high-density lipoprotein particle receptor binding / spherical high-density lipoprotein particle / positive regulation of hydrolase activity / negative regulation of response to cytokine stimulus / regulation of intestinal cholesterol absorption / protein oxidation / vitamin transport / cholesterol import / high-density lipoprotein particle binding /  ABC transporters in lipid homeostasis / blood vessel endothelial cell migration / negative regulation of heterotypic cell-cell adhesion / apolipoprotein receptor binding / negative regulation of cell adhesion molecule production / apolipoprotein A-I receptor binding / negative regulation of cytokine production involved in immune response / HDL assembly / peptidyl-methionine modification / negative regulation of very-low-density lipoprotein particle remodeling / phosphatidylcholine biosynthetic process / glucocorticoid metabolic process / phosphatidylcholine metabolic process / Chylomicron remodeling / phosphatidylcholine-sterol O-acyltransferase activator activity / positive regulation of phospholipid efflux / ABC transporters in lipid homeostasis / blood vessel endothelial cell migration / negative regulation of heterotypic cell-cell adhesion / apolipoprotein receptor binding / negative regulation of cell adhesion molecule production / apolipoprotein A-I receptor binding / negative regulation of cytokine production involved in immune response / HDL assembly / peptidyl-methionine modification / negative regulation of very-low-density lipoprotein particle remodeling / phosphatidylcholine biosynthetic process / glucocorticoid metabolic process / phosphatidylcholine metabolic process / Chylomicron remodeling / phosphatidylcholine-sterol O-acyltransferase activator activity / positive regulation of phospholipid efflux /  lipid storage / phospholipid homeostasis / lipid storage / phospholipid homeostasis /  Chylomicron assembly / positive regulation of cholesterol metabolic process / high-density lipoprotein particle clearance / phospholipid efflux / high-density lipoprotein particle remodeling / cholesterol transfer activity / Chylomicron assembly / positive regulation of cholesterol metabolic process / high-density lipoprotein particle clearance / phospholipid efflux / high-density lipoprotein particle remodeling / cholesterol transfer activity /  reverse cholesterol transport / reverse cholesterol transport /  chemorepellent activity / high-density lipoprotein particle assembly / very-low-density lipoprotein particle / cholesterol transport / positive regulation of CoA-transferase activity / lipoprotein biosynthetic process / high-density lipoprotein particle / endothelial cell proliferation / chemorepellent activity / high-density lipoprotein particle assembly / very-low-density lipoprotein particle / cholesterol transport / positive regulation of CoA-transferase activity / lipoprotein biosynthetic process / high-density lipoprotein particle / endothelial cell proliferation /  regulation of Cdc42 protein signal transduction / triglyceride homeostasis / HDL remodeling / negative regulation of interleukin-1 beta production / Scavenging by Class A Receptors / cholesterol efflux / regulation of Cdc42 protein signal transduction / triglyceride homeostasis / HDL remodeling / negative regulation of interleukin-1 beta production / Scavenging by Class A Receptors / cholesterol efflux /  cholesterol binding / positive regulation of Rho protein signal transduction / negative chemotaxis / adrenal gland development / cholesterol biosynthetic process / endocytic vesicle / positive regulation of cholesterol efflux / negative regulation of tumor necrosis factor-mediated signaling pathway / Scavenging of heme from plasma / positive regulation of phagocytosis / positive regulation of substrate adhesion-dependent cell spreading / Retinoid metabolism and transport / positive regulation of stress fiber assembly / endocytic vesicle lumen / cholesterol binding / positive regulation of Rho protein signal transduction / negative chemotaxis / adrenal gland development / cholesterol biosynthetic process / endocytic vesicle / positive regulation of cholesterol efflux / negative regulation of tumor necrosis factor-mediated signaling pathway / Scavenging of heme from plasma / positive regulation of phagocytosis / positive regulation of substrate adhesion-dependent cell spreading / Retinoid metabolism and transport / positive regulation of stress fiber assembly / endocytic vesicle lumen /  heat shock protein binding / cholesterol metabolic process / cholesterol homeostasis / integrin-mediated signaling pathway / heat shock protein binding / cholesterol metabolic process / cholesterol homeostasis / integrin-mediated signaling pathway /  Post-translational protein phosphorylation / Post-translational protein phosphorylation /  regulation of protein phosphorylation / regulation of protein phosphorylation /  phospholipid binding / Heme signaling / PPARA activates gene expression / negative regulation of inflammatory response / Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) / phospholipid binding / Heme signaling / PPARA activates gene expression / negative regulation of inflammatory response / Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) /  extracellular vesicle / Platelet degranulation / extracellular vesicle / Platelet degranulation /  amyloid-beta binding / cytoplasmic vesicle / collagen-containing extracellular matrix / secretory granule lumen / blood microparticle / protein stabilization / amyloid-beta binding / cytoplasmic vesicle / collagen-containing extracellular matrix / secretory granule lumen / blood microparticle / protein stabilization /  early endosome / G protein-coupled receptor signaling pathway / Amyloid fiber formation / early endosome / G protein-coupled receptor signaling pathway / Amyloid fiber formation /  endoplasmic reticulum lumen / endoplasmic reticulum lumen /  signaling receptor binding / signaling receptor binding /  enzyme binding / protein homodimerization activity / enzyme binding / protein homodimerization activity /  extracellular space / extracellular exosome / extracellular region / identical protein binding / extracellular space / extracellular exosome / extracellular region / identical protein binding /  plasma membrane / plasma membrane /  cytosol cytosolSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  SOLUTION SCATTERING / NUCLEAR REACTOR SOLUTION SCATTERING / NUCLEAR REACTOR | ||||||
 Authors Authors | Wu, Z. / Gogonea, V. / Lee, X. / Wagner, M.A. / Li, X.-M. / Huang, Y. / Undurti, A. / May, R.P. / Haertlein, M. / Moulin, M. ...Wu, Z. / Gogonea, V. / Lee, X. / Wagner, M.A. / Li, X.-M. / Huang, Y. / Undurti, A. / May, R.P. / Haertlein, M. / Moulin, M. / Gutsche, I. / Zaccai, G. / Didonato, J.A. / Hazen, L.S. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2009 Journal: J.Biol.Chem. / Year: 2009Title: Double superhelix model of high density lipoprotein. Authors: Wu, Z. / Gogonea, V. / Lee, X. / Wagner, M.A. / Li, X.M. / Huang, Y. / Undurti, A. / May, R.P. / Haertlein, M. / Moulin, M. / Gutsche, I. / Zaccai, G. / Didonato, J.A. / Hazen, S.L. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3k2s.cif.gz 3k2s.cif.gz | 289.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3k2s.ent.gz pdb3k2s.ent.gz | 293.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3k2s.json.gz 3k2s.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/k2/3k2s https://data.pdbj.org/pub/pdb/validation_reports/k2/3k2s ftp://data.pdbj.org/pub/pdb/validation_reports/k2/3k2s ftp://data.pdbj.org/pub/pdb/validation_reports/k2/3k2s | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein |  Apolipoprotein AI / Apo-AI / ApoA-I / Apolipoprotein A-I(1-242) Apolipoprotein AI / Apo-AI / ApoA-I / Apolipoprotein A-I(1-242)Mass: 28120.637 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Production host: Homo sapiens (human) / Production host:   Escherichia coli (E. coli) / References: UniProt: P02647 Escherichia coli (E. coli) / References: UniProt: P02647#2: Chemical | ChemComp-POV / (  POPC POPC#3: Chemical | ChemComp-CLR /  Cholesterol Cholesterol |
|---|
-Experimental details
-Experiment
| Experiment | Method:  SOLUTION SCATTERING / Number of used crystals: 1 SOLUTION SCATTERING / Number of used crystals: 1 |
|---|
-Data collection
| Diffraction | Mean temperature: 279 K | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source: NUCLEAR REACTOR / Type: OTHER / Wavelength: 5 Å | |||||||||||||||
| Detector | Detector: AREA DETECTOR / Date: Oct 15, 2008 | |||||||||||||||
| Radiation | Monochromator: GRAPHITE / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | |||||||||||||||
| Radiation wavelength | Wavelength : 5 Å / Relative weight: 1 : 5 Å / Relative weight: 1 | |||||||||||||||
| Reflection shell | Highest resolution: 5 Å | |||||||||||||||
| Soln scatter | Conc. range: 2-4 / Data analysis software list: PRIMUS, GNOM, DAMMIN / Data reduction software list: ILL IN-HOUSE PACKAGE / Detector type: AREA / Num. of time frames: 3 / Sample pH: 7.4 / Source beamline: NEUTRON CAMERA FOR REACTOR SOURCE / Source beamline instrument: D22 (LOQ) / Source class: N / Source type: ILL / Temperature: 301 K / Type: neutron
|
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement step | Cycle: LAST
| |||||||||||||||||||||||||||||||||||||||
| Soln scatter model | Method: CONSTRAINED SCATTERING FITTING OF HOMOLOGY MODELS Conformer selection criteria: THE MODELLED SCATTERING CURVES WERE ASSESSED BY CALCULATION OF THE RG IN THE SAME Q RANGES USED IN THE EXPERIMENTAL GUINIER FITS. MODELS WERE THEN RANKED USING A ...Conformer selection criteria: THE MODELLED SCATTERING CURVES WERE ASSESSED BY CALCULATION OF THE RG IN THE SAME Q RANGES USED IN THE EXPERIMENTAL GUINIER FITS. MODELS WERE THEN RANKED USING A GOODNESS-OF-FIT CHI SQUARE-FACTOR DEFINED BY THE DIFFERENCE BETWEEN THE CALCULATED AND EXPERIMENTAL SCATTERING INTENSITIES AND BASED ON THE EXPERIMENTAL CURVES IN THE Q RANGE EXTENDING TO 2.4 NM-1 (PROTEIN) AND 2.1 NM-1 (LIPID). Details: AN ALL ATOM COMPUTATIONAL MODEL WAS CONSTRUCTED BY COMBINING MOLELING WITH CONRAST VARIATION SANS, HYDROGEN-DEUTERIUM EXCHANGE (H/D-MS/MS), AND DISTANCE CONSTRAINTS FROM CROSS-LINKING, ...Details: AN ALL ATOM COMPUTATIONAL MODEL WAS CONSTRUCTED BY COMBINING MOLELING WITH CONRAST VARIATION SANS, HYDROGEN-DEUTERIUM EXCHANGE (H/D-MS/MS), AND DISTANCE CONSTRAINTS FROM CROSS-LINKING, FLUORESCENCE RESONANCE ENERGY TRANSFER AND ELECTRON SPIN RESONANCE. SANS LOW RESOLUTION STRUCTURES (12% AND 42% D2O) WERE USED AS SCAFFOLDS TO BUILD MOLECULAR MODELS FOR PROTEIN AND LIPID COMPONENTS OF NASCENT HDL. 256 MODELS WERE GENERATED AND ASSESSED USING GOODNESS-OF-FIT WITH BOTH SANS AND H/D-MS/MS DATA. THE NEUTRON SCATTERING CURVE I(Q) WAS CALCULATED ASSUMING ALL ATOM STRUCTURE, AND WITH CORRECTIONS FOR WAVELENGTH SPREAD AND BEAM DIVERGENCE. Entry fitting list: THE DOUBLE BELT MODEL (SEGREST JP, JBC 274, 31755 (1999)) Num. of conformers calculated: 256 / Num. of conformers submitted: 1 / Representative conformer: 1 Software author list: LINDAHL E, SALI A, DELANO WL, MERZEL F, RICHARDS RM, SVERGUN DI Software list: GROMACS, MODELLER, PYMOL, SASSIM, YALE:VOLUME/ACCESS, PRIMUS, GNOM, DAMMIN |
 Movie
Movie Controller
Controller












 PDBj
PDBj