-Search query
-Search result
Showing 1 - 50 of 644 items for (database: EMDB) & (Data entries: Updated only)

EMDB-40940: 
TRPV1 in Nanodisc bound with lysophosphatidic acid in all four monomers
Method: single particle / : Arnold WR, Cheng Y

EMDB-40941: 
TRPV1 in Nanodisc not bound with lysophosphatidic acid (apo)
Method: single particle / : Arnold WR, Cheng Y

EMDB-40949: 
TRPV1 in nanodisc bound with one LPA in one monomer
Method: single particle / : Arnold WR, Cheng Y

EMDB-40951: 
TRPV1 in nanodisc bound with two LPA molecules in opposite monomers
Method: single particle / : Arnold WR, Julius D, Cheng Y

EMDB-41005: 
TRPV1 in nanodisc bound with 2 LPA molecules in neighboring monomers
Method: single particle / : Arnold WR, Julius D, Cheng Y

EMDB-41006: 
TRPV1 in nanodisc bound with 3 LPA molecules
Method: single particle / : Arnold WR, Julius D, Cheng Y

EMDB-41847: 
TRPV1 in nanodisc bound with diC8-PIP2 in the dilated state
Method: single particle / : Arnold WR, Julius D, Cheng Y

EMDB-41848: 
TRPV1 in nanodisc bound with diC8-PIP2 in the closed state
Method: single particle / : Arnold WR, Julius D, Cheng Y

EMDB-41855: 
TRPV1 in nanodisc bound with PI-Br4 bound in Conformation 1 (monomer)
Method: single particle / : Arnold WR, Julius D, Cheng Y

EMDB-41857: 
TRPV1 in nanodisc bound with PI-Br4 bound in Conformation 2 (monomer)
Method: single particle / : Arnold WR, Julius D, Cheng Y

EMDB-41864: 
TRPV1 in nanodisc bound with empty vanilloid binding pocket at 4C
Method: single particle / : Arnold WR, Julius D, Cheng Y

EMDB-41866: 
TRPV1 in nanodisc bound with empty vanilloid binding pocket at 25C
Method: single particle / : Arnold WR, Julius D, Cheng Y

EMDB-41873: 
TRPV1 in nanodisc bound with PIP2-Br4
Method: single particle / : Arnold WR, Julius D, Cheng Y

EMDB-41879: 
TRPV1 in nanodisc bound with PI-Br4, consensus structure
Method: single particle / : Arnold WR, Julius D, Cheng Y

EMDB-17213: 
Human Mitochondrial Lon Y186F Mutant ADP Bound
Method: single particle / : Kereiche S, Bauer JA, Matyas P, Novacek J, Kutejova E

EMDB-17214: 
Human Mitochondrial Lon Y186E Mutant ADP Bound
Method: single particle / : Kereiche S, Bauer JA, Matyas P, Novacek J, Kutejova E

EMDB-43488: 
Cryo-EM structure of human invariant chain in complex with HLA-DR15
Method: single particle / : Wang N, Caveney NA, Jude KM, Garcia KC

EMDB-43501: 
Cryo-EM structure of human invariant chain in complex with HLA-DQ
Method: single particle / : Wang N, Caveney NA, Jude KM, Garcia KC

EMDB-42383: 
Cryo-EM map of the human CTF18-RFC-PCNA binary complex in the three-subunit binding state (state 2)
Method: single particle / : Wang F, He Q, Li H

EMDB-42384: 
Cryo-EM map of the human CTF18-RFC-PCNA binary complex in the four-subunit binding state (state 3)
Method: single particle / : Wang F, He Q, Li H

EMDB-42385: 
Cryo-EM map of the human CTF18-RFC-PCNA-DNA ternary complex with narrow PCNA opening state I (state 5)
Method: single particle / : Wang F, He Q, Li H

EMDB-42386: 
Cryo-EM map of the human CTF18-RFC-PCNA-DNA ternary complex in the five-subunit binding state (state 4)
Method: single particle / : Wang F, He Q, Li H

EMDB-42388: 
Cryo-EM map of the human CTF18-RFC-PCNA-DNA ternary complex with narrow PCNA opening state II (state 6)
Method: single particle / : Wang F, He Q, Li H

EMDB-42389: 
Cryo-EM map of the human CTF18-RFC-PCNA-DNA ternary complex with cracked and closed PCNA (state 7)
Method: single particle / : Wang F, He Q, Li H

EMDB-42406: 
Cryo-EM map of the human CTF18-RFC alone in the apo state (State 1)
Method: single particle / : Wang F, He Q, Li H

EMDB-17216: 
CRYO-EM STRUCTURE OF LEISHMANIA MAJOR 80S RIBOSOME : PARENTAL STRAIN
Method: single particle / : Rajan KS, Yonath A

EMDB-43683: 
Cryo-EM structure of FLVCR2 in the inward-facing state with choline bound
Method: single particle / : Cater RJ, Mancia F

EMDB-43684: 
Cryo-EM structure of FLVCR2 in the outward-facing state with choline bound
Method: single particle / : Cater RJ, Mancia F

EMDB-40681: 
SpRY-Cas9:gRNA complex targeting TGG PAM DNA
Method: single particle / : Hibshman GN, Bravo JPK, Taylor DW
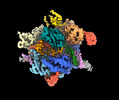
EMDB-40705: 
SpRY-Cas9:gRNA complex targeting TTC PAM DNA
Method: single particle / : Hibshman GN, Bravo JPK, Taylor DW

EMDB-40740: 
SpRY-Cas9:gRNA complex targeting TAC PAM DNA
Method: single particle / : Hibshman GN, Bravo JPK, Taylor DW

EMDB-41073: 
SpRY-Cas9:gRNA complex targeting TAC PAM DNA with 0 bp R-loop
Method: single particle / : Hibshman GN, Bravo JPK, Taylor DW
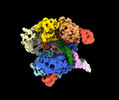
EMDB-41074: 
SpRY-Cas9:gRNA complex targeting TAC PAM DNA with 2 bp R-loop
Method: single particle / : Hibshman GN, Bravo JPK, Taylor DW

EMDB-41079: 
SpRY-Cas9:gRNA complex targeting TAC PAM DNA with 10 bp R-loop
Method: single particle / : Hibshman GN, Bravo JPK, Taylor DW

EMDB-41080: 
SpRY-Cas9:gRNA complex targeting TAC PAM DNA with 13 bp R-loop
Method: single particle / : Hibshman GN, Bravo JPK, Taylor DW

EMDB-41083: 
SpRY-Cas9:gRNA complex targeting TAC PAM DNA with 18 bp R-loop
Method: single particle / : Hibshman GN, Bravo JPK, Taylor DW

EMDB-41084: 
SpRY-Cas9:gRNA complex bound to non-target DNA with 1 bp R-loop
Method: single particle / : Hibshman GN, Bravo JPK, Taylor DW

EMDB-41085: 
SpRY-Cas9:gRNA complex targeting TAC PAM DNA with 3 bp R-loop
Method: single particle / : Hibshman GN, Bravo JPK, Taylor DW
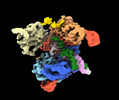
EMDB-41086: 
SpRY-Cas9:gRNA complex bound to non-target DNA with 6 bp R-loop
Method: single particle / : Hibshman GN, Bravo JPK, Taylor DW

EMDB-41087: 
SpRY-Cas9:gRNA complex bound to non-target DNA with 8 bp R-loop
Method: single particle / : Hibshman GN, Bravo JPK, Taylor DW
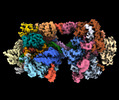
EMDB-41093: 
SpRYmer bound to NAC PAM DNA
Method: single particle / : Hibshman GN, Bravo JPK, Taylor DW

EMDB-41775: 
SpG Cas9 with NGC PAM DNA target
Method: single particle / : Bravo JPK, Hibshman GN, Taylor DW

EMDB-41867: 
SpG Cas9 with NGG PAM DNA target
Method: single particle / : Bravo JPK, Hibshman GN, Taylor DW

EMDB-17189: 
Structure of the human neutral amino acid transporter ASCT2 in complex with nanobody 469
Method: single particle / : Canul-Tec J, Reyes N

EMDB-17192: 
Complex of human ASCT2 with Syncytin-1
Method: single particle / : Khare S, Reyes N

EMDB-17193: 
Complex of ASCT2 with Suppressyn
Method: single particle / : Khare S, Kumar A, Reyes N

EMDB-17194: 
Heterotrimeric Complex of Human ASCT2 with Syncytin-1
Method: single particle / : Khare S, Reyes N

EMDB-18879: 
Roco protein from C. tepidum in the GTP state bound to an activating Nanobody
Method: single particle / : Galicia C, Fislage M, Versees W

EMDB-18882: 
Roco protein from C. tepidum in the GTP state bound to the activating Nanobodies NbRoco1 and NbRoco2
Method: single particle / : Galicia C, Fislage M, Versees W

EMDB-18885: 
Focused map on the LRR domain of the Roco protein from C. tepidum bound to the activating Nanobody NbRoco2
Method: single particle / : Galicia C, Versees W
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
