+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8jjr | ||||||
|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Symbiodinium photosystem I | ||||||
 Components Components |
| ||||||
 Keywords Keywords |  PHOTOSYNTHESIS / Symbiodinium / PHOTOSYNTHESIS / Symbiodinium /  Photosystem I Photosystem I | ||||||
| Function / homology |  Function and homology information Function and homology information BETA-CAROTENE / BETA-CAROTENE /  CHLOROPHYLL A / Chem-DD6 / DIGALACTOSYL DIACYL GLYCEROL (DGDG) / Chlorophyll c2 / 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE / 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE / CHLOROPHYLL A / Chem-DD6 / DIGALACTOSYL DIACYL GLYCEROL (DGDG) / Chlorophyll c2 / 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE / 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE /  PERIDININ / PERIDININ /  PHYLLOQUINONE / PHYLLOQUINONE /  IRON/SULFUR CLUSTER ... IRON/SULFUR CLUSTER ... BETA-CAROTENE / BETA-CAROTENE /  CHLOROPHYLL A / Chem-DD6 / DIGALACTOSYL DIACYL GLYCEROL (DGDG) / Chlorophyll c2 / 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE / 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE / CHLOROPHYLL A / Chem-DD6 / DIGALACTOSYL DIACYL GLYCEROL (DGDG) / Chlorophyll c2 / 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE / 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE /  PERIDININ / PERIDININ /  PHYLLOQUINONE / PHYLLOQUINONE /  IRON/SULFUR CLUSTER / Chem-SQD / : IRON/SULFUR CLUSTER / Chem-SQD / : Similarity search - Component | ||||||
| Biological species |  Symbiodinium sp. (eukaryote) Symbiodinium sp. (eukaryote) | ||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.8 Å cryo EM / Resolution: 2.8 Å | ||||||
 Authors Authors | Zhao, L.S. / Wang, N. / Li, K. / Zhang, Y.Z. / Liu, L.N. | ||||||
| Funding support |  China, 1items China, 1items
| ||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Architecture of symbiotic dinoflagellate photosystem I-light-harvesting supercomplex in Symbiodinium. Authors: Long-Sheng Zhao / Ning Wang / Kang Li / Chun-Yang Li / Jian-Ping Guo / Fei-Yu He / Gui-Ming Liu / Xiu-Lan Chen / Jun Gao / Lu-Ning Liu / Yu-Zhong Zhang /   Abstract: Symbiodinium are the photosynthetic endosymbionts for corals and play a vital role in supplying their coral hosts with photosynthetic products, forming the nutritional foundation for high-yield coral ...Symbiodinium are the photosynthetic endosymbionts for corals and play a vital role in supplying their coral hosts with photosynthetic products, forming the nutritional foundation for high-yield coral reef ecosystems. Here, we determine the cryo-electron microscopy structure of Symbiodinium photosystem I (PSI) supercomplex with a PSI core composed of 13 subunits including 2 previously unidentified subunits, PsaT and PsaU, as well as 13 peridinin-Chl a/c-binding light-harvesting antenna proteins (AcpPCIs). The PSI-AcpPCI supercomplex exhibits distinctive structural features compared to their red lineage counterparts, including extended termini of PsaD/E/I/J/L/M/R and AcpPCI-1/3/5/7/8/11 subunits, conformational changes in the surface loops of PsaA and PsaB subunits, facilitating the association between the PSI core and peripheral antennae. Structural analysis and computational calculation of excitation energy transfer rates unravel specific pigment networks in Symbiodinium PSI-AcpPCI for efficient excitation energy transfer. Overall, this study provides a structural basis for deciphering the mechanisms governing light harvesting and energy transfer in Symbiodinium PSI-AcpPCI supercomplexes adapted to their symbiotic ecosystem, as well as insights into the evolutionary diversity of PSI-LHCI among various photosynthetic organisms. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8jjr.cif.gz 8jjr.cif.gz | 1.2 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8jjr.ent.gz pdb8jjr.ent.gz | 1.1 MB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8jjr.json.gz 8jjr.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/jj/8jjr https://data.pdbj.org/pub/pdb/validation_reports/jj/8jjr ftp://data.pdbj.org/pub/pdb/validation_reports/jj/8jjr ftp://data.pdbj.org/pub/pdb/validation_reports/jj/8jjr | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  36366MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
+Protein , 26 types, 26 molecules ABCDEFGHIJKTUabcdefijlmrxy
-Sugars , 2 types, 5 molecules 
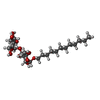

| #35: Sugar | ChemComp-DGD / #36: Sugar | ChemComp-LMU / | |
|---|
-Non-polymers , 11 types, 318 molecules 


















| #27: Chemical | ChemComp-CLA /  Chlorophyll a Chlorophyll a#28: Chemical | ChemComp-KC2 /  Chlorophyll c Chlorophyll c#29: Chemical | ChemComp-UIX / [( Mass: 642.907 Da / Num. of mol.: 16 / Source method: obtained synthetically / Formula: C42H58O5 / Feature type: SUBJECT OF INVESTIGATION #30: Chemical | ChemComp-DD6 / (  Diadinoxanthin Diadinoxanthin#31: Chemical | #32: Chemical | ChemComp-PID /  Peridinin Peridinin#33: Chemical | ChemComp-LMG / #34: Chemical | ChemComp-LHG /  Phosphatidylglycerol Phosphatidylglycerol#37: Chemical |  Phytomenadione Phytomenadione#38: Chemical | ChemComp-BCR /  Β-Carotene Β-Carotene#39: Chemical |  Iron–sulfur cluster Iron–sulfur cluster |
|---|
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method:  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Photosystem I of Symbiodinium / Type: COMPLEX / Entity ID: #1-#26 / Source: NATURAL / Type: COMPLEX / Entity ID: #1-#26 / Source: NATURAL |
|---|---|
| Molecular weight | Experimental value: NO |
| Source (natural) | Organism:  Symbiodinium sp. (eukaryote) Symbiodinium sp. (eukaryote) |
| Buffer solution | pH: 6.5 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied : NO / Vitrification applied : NO / Vitrification applied : YES : YES |
Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source : :  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: SPOT SCAN FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: SPOT SCAN |
| Electron lens | Mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 1800 nm / Nominal defocus min: 1000 nm Bright-field microscopy / Nominal defocus max: 1800 nm / Nominal defocus min: 1000 nm |
| Image recording | Electron dose: 50 e/Å2 / Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) |
- Processing
Processing
CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
3D reconstruction | Resolution: 2.8 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 161863 / Symmetry type: POINT | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller



 PDBj
PDBj
















