+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8i2z | ||||||
|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the zeaxanthin-bound kin4B8 | ||||||
 Components Components | Xanthorhodopsin | ||||||
 Keywords Keywords |  PROTON TRANSPORT / PROTON TRANSPORT /  Rhodopsin / Rhodopsin /  Cryo-EM / anntena Cryo-EM / anntena | ||||||
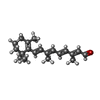
| Function / homology | Zeaxanthin /  RETINAL / Xanthorhodopsin RETINAL / Xanthorhodopsin Function and homology information Function and homology information | ||||||
| Biological species |  uncultured Bdellovibrionales bacterium (environmental samples) uncultured Bdellovibrionales bacterium (environmental samples) | ||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.3 Å cryo EM / Resolution: 2.3 Å | ||||||
 Authors Authors | Murakoshi, S. / Chazan, A. / Shihoya, W. / Beja, O. / Nureki, O. | ||||||
| Funding support |  Japan, 1items Japan, 1items
| ||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: Phototrophy by antenna-containing rhodopsin pumps in aquatic environments. Authors: Ariel Chazan / Ishita Das / Takayoshi Fujiwara / Shunya Murakoshi / Andrey Rozenberg / Ana Molina-Márquez / Fumiya K Sano / Tatsuki Tanaka / Patricia Gómez-Villegas / Shirley Larom / Alina ...Authors: Ariel Chazan / Ishita Das / Takayoshi Fujiwara / Shunya Murakoshi / Andrey Rozenberg / Ana Molina-Márquez / Fumiya K Sano / Tatsuki Tanaka / Patricia Gómez-Villegas / Shirley Larom / Alina Pushkarev / Partha Malakar / Masumi Hasegawa / Yuya Tsukamoto / Tomohiro Ishizuka / Masae Konno / Takashi Nagata / Yosuke Mizuno / Kota Katayama / Rei Abe-Yoshizumi / Sanford Ruhman / Keiichi Inoue / Hideki Kandori / Rosa León / Wataru Shihoya / Susumu Yoshizawa / Mordechai Sheves / Osamu Nureki / Oded Béjà /     Abstract: Energy transfer from light-harvesting ketocarotenoids to the light-driven proton pump xanthorhodopsins has been previously demonstrated in two unique cases: an extreme halophilic bacterium and a ...Energy transfer from light-harvesting ketocarotenoids to the light-driven proton pump xanthorhodopsins has been previously demonstrated in two unique cases: an extreme halophilic bacterium and a terrestrial cyanobacterium. Attempts to find carotenoids that bind and transfer energy to abundant rhodopsin proton pumps from marine photoheterotrophs have thus far failed. Here we detected light energy transfer from the widespread hydroxylated carotenoids zeaxanthin and lutein to the retinal moiety of xanthorhodopsins and proteorhodopsins using functional metagenomics combined with chromophore extraction from the environment. The light-harvesting carotenoids transfer up to 42% of the harvested energy in the violet- or blue-light range to the green-light absorbing retinal chromophore. Our data suggest that these antennas may have a substantial effect on rhodopsin phototrophy in the world's lakes, seas and oceans. However, the functional implications of our findings are yet to be discovered. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8i2z.cif.gz 8i2z.cif.gz | 61.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8i2z.ent.gz pdb8i2z.ent.gz | 44.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8i2z.json.gz 8i2z.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/i2/8i2z https://data.pdbj.org/pub/pdb/validation_reports/i2/8i2z ftp://data.pdbj.org/pub/pdb/validation_reports/i2/8i2z ftp://data.pdbj.org/pub/pdb/validation_reports/i2/8i2z | HTTPS FTP |
|---|
-Related structure data
| Related structure data | 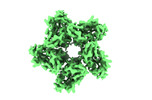 35143MC  7ytbC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 | x 5
|
| 2 |
|
| 3 | 
|
| Symmetry | Point symmetry: (Schoenflies symbol : C5 (5 fold cyclic : C5 (5 fold cyclic )) )) |
- Components
Components
| #1: Protein | Mass: 28745.842 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  uncultured Bdellovibrionales bacterium (environmental samples) uncultured Bdellovibrionales bacterium (environmental samples)Gene: tmp_000014 / Production host:   Escherichia coli (E. coli) / References: UniProt: A0A977XLG0 Escherichia coli (E. coli) / References: UniProt: A0A977XLG0 |
|---|---|
| #2: Chemical | ChemComp-RET /  Retinal Retinal |
| #3: Chemical | ChemComp-K3I / |
| #4: Water | ChemComp-HOH /  Water Water |
| Has ligand of interest | Y |
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method:  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Kin4B8 / Type: ORGANELLE OR CELLULAR COMPONENT / Entity ID: #1 / Source: RECOMBINANT | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight | Experimental value: NO | |||||||||||||||
| Source (natural) | Organism:  uncultured Bdellovibrionales bacterium (environmental samples) uncultured Bdellovibrionales bacterium (environmental samples) | |||||||||||||||
| Source (recombinant) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) | |||||||||||||||
| Buffer solution | pH: 8 / Details: 150 mM NaCl, 20 mM Tris-HCl | |||||||||||||||
| Buffer component |
| |||||||||||||||
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied : NO / Vitrification applied : NO / Vitrification applied : YES : YES | |||||||||||||||
| Specimen support | Grid material: GOLD / Grid mesh size: 300 divisions/in. / Grid type: Quantifoil R1.2/1.3 | |||||||||||||||
Vitrification | Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 277 K / Details: Vitrification carried out in ethane atmosphere. |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: TFS KRIOS |
| Electron gun | Electron source : :  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 1600 nm / Nominal defocus min: 600 nm Bright-field microscopy / Nominal defocus max: 1600 nm / Nominal defocus min: 600 nm |
| Image recording | Electron dose: 50.14 e/Å2 / Film or detector model: GATAN K3 (6k x 4k) |
- Processing
Processing
| Software | Name: REFMAC / Version: 5.8.0267 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
3D reconstruction | Resolution: 2.3 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 4337540 / Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | Resolution: 2→107.9 Å / Cor.coef. Fo:Fc: 0.899 / SU B: 3.934 / SU ML: 0.087 / ESU R: 0.05 Stereochemistry target values: MAXIMUM LIKELIHOOD WITH PHASES Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: PARAMETERS FOR MASK CACLULATION | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 86.425 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: 1 / Total: 2050 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller



 PDBj
PDBj