+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8fv5 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
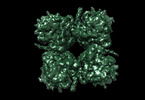
| Title | Representation of 16-mer phiPA3 PhuN Lattice, p2 | |||||||||
 Components Components | Maltose/maltodextrin-binding periplasmic protein, phiPA3 PhuN | |||||||||
 Keywords Keywords |  VIRAL PROTEIN / phiPA3 protein / shell protein / PhuN / phage nucleus VIRAL PROTEIN / phiPA3 protein / shell protein / PhuN / phage nucleus | |||||||||
| Function / homology |  Function and homology information Function and homology informationdetection of maltose stimulus / maltose transport complex /  maltose binding / maltose transport / maltodextrin transmembrane transport / carbohydrate transport / carbohydrate transmembrane transporter activity / ATP-binding cassette (ABC) transporter complex, substrate-binding subunit-containing / ATP-binding cassette (ABC) transporter complex / cell chemotaxis ...detection of maltose stimulus / maltose transport complex / maltose binding / maltose transport / maltodextrin transmembrane transport / carbohydrate transport / carbohydrate transmembrane transporter activity / ATP-binding cassette (ABC) transporter complex, substrate-binding subunit-containing / ATP-binding cassette (ABC) transporter complex / cell chemotaxis ...detection of maltose stimulus / maltose transport complex /  maltose binding / maltose transport / maltodextrin transmembrane transport / carbohydrate transport / carbohydrate transmembrane transporter activity / ATP-binding cassette (ABC) transporter complex, substrate-binding subunit-containing / ATP-binding cassette (ABC) transporter complex / cell chemotaxis / outer membrane-bounded periplasmic space / host cell cytoplasm / maltose binding / maltose transport / maltodextrin transmembrane transport / carbohydrate transport / carbohydrate transmembrane transporter activity / ATP-binding cassette (ABC) transporter complex, substrate-binding subunit-containing / ATP-binding cassette (ABC) transporter complex / cell chemotaxis / outer membrane-bounded periplasmic space / host cell cytoplasm /  periplasmic space / DNA damage response / periplasmic space / DNA damage response /  membrane membraneSimilarity search - Function | |||||||||
| Biological species |   Escherichia coli (E. coli) Escherichia coli (E. coli) Pseudomonas phage PhiPA3 (virus) Pseudomonas phage PhiPA3 (virus) | |||||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 4.21 Å cryo EM / Resolution: 4.21 Å | |||||||||
 Authors Authors | Nieweglowska, E.S. / Brilot, A.F. / Mendez-Moran, M. / Kokontis, C. / Baek, M. / Li, J. / Cheng, Y. / Baker, D. / Bondy-Denomy, J. / Agard, D.A. | |||||||||
| Funding support |  United States, 2items United States, 2items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: The ϕPA3 phage nucleus is enclosed by a self-assembling 2D crystalline lattice. Authors: Eliza S Nieweglowska / Axel F Brilot / Melissa Méndez-Moran / Claire Kokontis / Minkyung Baek / Junrui Li / Yifan Cheng / David Baker / Joseph Bondy-Denomy / David A Agard /  Abstract: To protect themselves from host attack, numerous jumbo bacteriophages establish a phage nucleus-a micron-scale, proteinaceous structure encompassing the replicating phage DNA. Bacteriophage and host ...To protect themselves from host attack, numerous jumbo bacteriophages establish a phage nucleus-a micron-scale, proteinaceous structure encompassing the replicating phage DNA. Bacteriophage and host proteins associated with replication and transcription are concentrated inside the phage nucleus while other phage and host proteins are excluded, including CRISPR-Cas and restriction endonuclease host defense systems. Here, we show that nucleus fragments isolated from ϕPA3 infected Pseudomonas aeruginosa form a 2-dimensional lattice, having p2 or p4 symmetry. We further demonstrate that recombinantly purified primary Phage Nuclear Enclosure (PhuN) protein spontaneously assembles into similar 2D sheets with p2 and p4 symmetry. We resolve the dominant p2 symmetric state to 3.9 Å by cryo-EM. Our structure reveals a two-domain core, organized into quasi-symmetric tetramers. Flexible loops and termini mediate adaptable inter-tetramer contacts that drive subunit assembly into a lattice and enable the adoption of different symmetric states. While the interfaces between subunits are mostly well packed, two are open, forming channels that likely have functional implications for the transport of proteins, mRNA, and small molecules. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8fv5.cif.gz 8fv5.cif.gz | 3.1 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8fv5.ent.gz pdb8fv5.ent.gz | Display |  PDB format PDB format | |
| PDBx/mmJSON format |  8fv5.json.gz 8fv5.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/fv/8fv5 https://data.pdbj.org/pub/pdb/validation_reports/fv/8fv5 ftp://data.pdbj.org/pub/pdb/validation_reports/fv/8fv5 ftp://data.pdbj.org/pub/pdb/validation_reports/fv/8fv5 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  29451MC  8fneC C: citing same article ( M: map data used to model this data |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein | Mass: 110126.078 Da / Num. of mol.: 32 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Escherichia coli (strain K12) (bacteria), (gene. exp.) Escherichia coli (strain K12) (bacteria), (gene. exp.)  Pseudomonas phage PhiPA3 (virus) Pseudomonas phage PhiPA3 (virus)Strain: K12 / Gene: malE, 053 / Production host:   Escherichia coli (E. coli) / Strain (production host): BL21 Star (DE3) pLysS / References: UniProt: P0AEX9, UniProt: F8SJT5 Escherichia coli (E. coli) / Strain (production host): BL21 Star (DE3) pLysS / References: UniProt: P0AEX9, UniProt: F8SJT5 |
|---|
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: 2D ARRAY / 3D reconstruction method:  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Lattice of phiPA3 PhuN Tetramer, p2 / Type: COMPLEX / Entity ID: all / Source: MULTIPLE SOURCES | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight | Experimental value: NO | ||||||||||||||||||||||||||||
| Source (natural) | Organism:  Pseudomonas phage PhiPA3 (virus) Pseudomonas phage PhiPA3 (virus) | ||||||||||||||||||||||||||||
| Source (recombinant) | Organism:   Escherichia coli (E. coli) / Strain: BL21 Star (DE3) pLysS Escherichia coli (E. coli) / Strain: BL21 Star (DE3) pLysS | ||||||||||||||||||||||||||||
| Buffer solution | pH: 6.5 Details: 0.25 cOmplete Protease Inhibitor Tablet also included | ||||||||||||||||||||||||||||
| Buffer component |
| ||||||||||||||||||||||||||||
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied : NO / Vitrification applied : NO / Vitrification applied : YES : YES | ||||||||||||||||||||||||||||
Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 283.15 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source : :  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD Bright-field microscopy / Nominal magnification: 105000 X / Nominal defocus max: 2500 nm / Nominal defocus min: 800 nm / Cs Bright-field microscopy / Nominal magnification: 105000 X / Nominal defocus max: 2500 nm / Nominal defocus min: 800 nm / Cs : 2.7 mm : 2.7 mm |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Electron dose: 67 e/Å2 / Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) |
- Processing
Processing
| Software | Name: UCSF ChimeraX / Version: 1.5/v9 / Classification: model building / URL: https://www.rbvi.ucsf.edu/chimerax/ / Os: macOS / Type: package | |||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software |
| |||||||||||||||||||||||||||||||||||||||||||||
CTF correction | Type: NONE | |||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Point symmetry : C2 (2 fold cyclic : C2 (2 fold cyclic ) ) | |||||||||||||||||||||||||||||||||||||||||||||
3D reconstruction | Resolution: 4.21 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 161270 / Symmetry type: POINT | |||||||||||||||||||||||||||||||||||||||||||||
| Atomic model building | Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller




 PDBj
PDBj


