[English] 日本語
 Yorodumi
Yorodumi- PDB-8fiy: Cryo-EM structure of E. coli RNA polymerase Elongation complex in... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8fiy | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of E. coli RNA polymerase Elongation complex in the Transcription-Translation Complex (RNAP in an anti-swiveled conformation) | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | TRANSCRIPTION/DNA/RNA /  RNA polymerase / RNA polymerase /  ribosome / ribosome /  coupling / coupling /  TRANSCRIPTION / TRANSCRIPTION-DNA-RNA complex TRANSCRIPTION / TRANSCRIPTION-DNA-RNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationsubmerged biofilm formation / cellular response to cell envelope stress / cytosolic DNA-directed RNA polymerase complex / regulation of DNA-templated transcription initiation / bacterial-type flagellum assembly / bacterial-type flagellum-dependent cell motility / nitrate assimilation / transcription elongation factor complex / regulation of DNA-templated transcription elongation / transcription antitermination ...submerged biofilm formation / cellular response to cell envelope stress / cytosolic DNA-directed RNA polymerase complex / regulation of DNA-templated transcription initiation / bacterial-type flagellum assembly / bacterial-type flagellum-dependent cell motility / nitrate assimilation / transcription elongation factor complex / regulation of DNA-templated transcription elongation / transcription antitermination /  cell motility / DNA-templated transcription initiation / cell motility / DNA-templated transcription initiation /  ribonucleoside binding / DNA-directed 5'-3' RNA polymerase activity / ribonucleoside binding / DNA-directed 5'-3' RNA polymerase activity /  DNA-directed RNA polymerase / response to heat / intracellular iron ion homeostasis / DNA-directed RNA polymerase / response to heat / intracellular iron ion homeostasis /  protein dimerization activity / response to antibiotic / magnesium ion binding / protein dimerization activity / response to antibiotic / magnesium ion binding /  DNA binding / zinc ion binding / DNA binding / zinc ion binding /  membrane / membrane /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Escherichia coli K-12 (bacteria) Escherichia coli K-12 (bacteria)  Escherichia phage Lambda (virus) Escherichia phage Lambda (virus) | |||||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 7.3 Å cryo EM / Resolution: 7.3 Å | |||||||||
 Authors Authors | Florez Ariza, A. / Wee, L. / Tong, A. / Canari, C. / Grob, P. / Nogales, E. / Bustamante, C. | |||||||||
| Funding support |  United States, 2items United States, 2items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2023 Journal: Cell / Year: 2023Title: A trailing ribosome speeds up RNA polymerase at the expense of transcript fidelity via force and allostery. Authors: Liang Meng Wee / Alexander B Tong / Alfredo Jose Florez Ariza / Cristhian Cañari-Chumpitaz / Patricia Grob / Eva Nogales / Carlos J Bustamante /  Abstract: In prokaryotes, translation can occur on mRNA that is being transcribed in a process called coupling. How the ribosome affects the RNA polymerase (RNAP) during coupling is not well understood. Here, ...In prokaryotes, translation can occur on mRNA that is being transcribed in a process called coupling. How the ribosome affects the RNA polymerase (RNAP) during coupling is not well understood. Here, we reconstituted the E. coli coupling system and demonstrated that the ribosome can prevent pausing and termination of RNAP and double the overall transcription rate at the expense of fidelity. Moreover, we monitored single RNAPs coupled to ribosomes and show that coupling increases the pause-free velocity of the polymerase and that a mechanical assisting force is sufficient to explain the majority of the effects of coupling. Also, by cryo-EM, we observed that RNAPs with a terminal mismatch adopt a backtracked conformation, while a coupled ribosome allosterically induces these polymerases toward a catalytically active anti-swiveled state. Finally, we demonstrate that prolonged RNAP pausing is detrimental to cell viability, which could be prevented by polymerase reactivation through a coupled ribosome. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8fiy.cif.gz 8fiy.cif.gz | 568.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8fiy.ent.gz pdb8fiy.ent.gz | 460.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8fiy.json.gz 8fiy.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/fi/8fiy https://data.pdbj.org/pub/pdb/validation_reports/fi/8fiy ftp://data.pdbj.org/pub/pdb/validation_reports/fi/8fiy ftp://data.pdbj.org/pub/pdb/validation_reports/fi/8fiy | HTTPS FTP |
|---|
-Related structure data
| Related structure data | 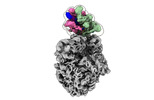 29213MC  8fixC  8fizC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-DNA-directed RNA polymerase subunit ... , 3 types, 4 molecules ABCD
| #1: Protein |  Polymerase / RNAP subunit alpha / RNA polymerase subunit alpha / Transcriptase subunit alpha Polymerase / RNAP subunit alpha / RNA polymerase subunit alpha / Transcriptase subunit alphaMass: 36558.680 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Escherichia coli K-12 (bacteria) / Gene: rpoA, pez, phs, sez, b3295, JW3257 / Production host: Escherichia coli K-12 (bacteria) / Gene: rpoA, pez, phs, sez, b3295, JW3257 / Production host:   Escherichia coli BL21 (bacteria) / References: UniProt: P0A7Z4, Escherichia coli BL21 (bacteria) / References: UniProt: P0A7Z4,  DNA-directed RNA polymerase DNA-directed RNA polymerase#2: Protein | |  Polymerase / RNAP subunit beta / RNA polymerase subunit beta / Transcriptase subunit beta Polymerase / RNAP subunit beta / RNA polymerase subunit beta / Transcriptase subunit betaMass: 150820.875 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Escherichia coli K-12 (bacteria) Escherichia coli K-12 (bacteria)Gene: rpoB, groN, nitB, rif, ron, stl, stv, tabD, b3987, JW3950 Production host:   Escherichia coli BL21 (bacteria) / References: UniProt: P0A8V2, Escherichia coli BL21 (bacteria) / References: UniProt: P0A8V2,  DNA-directed RNA polymerase DNA-directed RNA polymerase#3: Protein | |  Polymerase / RNAP subunit beta' / RNA polymerase subunit beta' / Transcriptase subunit beta' Polymerase / RNAP subunit beta' / RNA polymerase subunit beta' / Transcriptase subunit beta'Mass: 155366.781 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Escherichia coli K-12 (bacteria) / Gene: rpoC, tabB, b3988, JW3951 / Production host: Escherichia coli K-12 (bacteria) / Gene: rpoC, tabB, b3988, JW3951 / Production host:   Escherichia coli BL21 (bacteria) / References: UniProt: P0A8T7, Escherichia coli BL21 (bacteria) / References: UniProt: P0A8T7,  DNA-directed RNA polymerase DNA-directed RNA polymerase |
|---|
-DNA chain , 2 types, 2 molecules NT
| #4: DNA chain | Mass: 4497.951 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.)   Escherichia phage Lambda (virus) Escherichia phage Lambda (virus) |
|---|---|
| #5: DNA chain | Mass: 7158.604 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.)   Escherichia phage Lambda (virus) Escherichia phage Lambda (virus) |
-RNA chain , 1 types, 1 molecules R
| #6: RNA chain |  Mass: 2854.782 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.)   Escherichia phage Lambda (virus) Escherichia phage Lambda (virus) |
|---|
-Non-polymers , 2 types, 3 molecules 


| #7: Chemical | | #8: Chemical | ChemComp-MG / | |
|---|
-Details
| Has ligand of interest | N |
|---|
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method:  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Cryo-EM structure of E. coli RNA polymerase Elongation complex (in the Transcription-Translation Complex) harboring a terminal mismatch Type: COMPLEX / Entity ID: #1-#6 / Source: MULTIPLE SOURCES |
|---|---|
| Molecular weight | Value: 0.4 MDa / Experimental value: NO |
| Buffer solution | pH: 7.5 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied : NO / Vitrification applied : NO / Vitrification applied : YES : YES |
| Specimen support | Grid type: C-flat-1.2/1.3 |
Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TALOS ARCTICA |
| Electron gun | Electron source : :  FIELD EMISSION GUN / Accelerating voltage: 200 kV / Illumination mode: OTHER FIELD EMISSION GUN / Accelerating voltage: 200 kV / Illumination mode: OTHER |
| Electron lens | Mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2500 nm / Nominal defocus min: 1000 nm Bright-field microscopy / Nominal defocus max: 2500 nm / Nominal defocus min: 1000 nm |
| Image recording | Electron dose: 50 e/Å2 / Film or detector model: GATAN K3 (6k x 4k) |
- Processing
Processing
| Software | Name: PHENIX / Version: 1.18.2_3874: / Classification: refinement | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||
| Symmetry | Point symmetry : C1 (asymmetric) : C1 (asymmetric) | ||||||||||||||||||||||||
3D reconstruction | Resolution: 7.3 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 18629 / Symmetry type: POINT | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller




 PDBj
PDBj









































