[English] 日本語
 Yorodumi
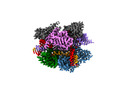
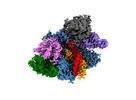
Yorodumi- PDB-8d2o: Structure of Acidothermus cellulolyticus Cas9 ternary complex (Po... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8d2o | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Acidothermus cellulolyticus Cas9 ternary complex (Post-cleavage 2) | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | RNA BINDING PROTEIN/DNA/RNA /  Cas9 / AcCas9 / Cas9 / AcCas9 /  Crispr / Post-cleavage 2 / Crispr / Post-cleavage 2 /  RNA BINDING PROTEIN / RNA BINDING PROTEIN-DNA-RNA complex RNA BINDING PROTEIN / RNA BINDING PROTEIN-DNA-RNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology information endonuclease activity / defense response to virus / endonuclease activity / defense response to virus /  DNA binding / DNA binding /  RNA binding / zinc ion binding RNA binding / zinc ion bindingSimilarity search - Function | |||||||||
| Biological species |   Acidothermus cellulolyticus 11B (bacteria) Acidothermus cellulolyticus 11B (bacteria)synthetic construct (others) | |||||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.66 Å cryo EM / Resolution: 2.66 Å | |||||||||
 Authors Authors | Rai, J. / Das, A. / Li, H. | |||||||||
| Funding support |  United States, 2items United States, 2items
| |||||||||
 Citation Citation |  Journal: Nat Catal / Year: 2023 Journal: Nat Catal / Year: 2023Title: Coupled catalytic states and the role of metal coordination in Cas9. Authors: Anuska Das / Jay Rai / Mitchell O Roth / Yuerong Shu / Megan L Medina / Mackenzie R Barakat / Hong Li /  Abstract: Controlling the activity of the CRISPR-Cas9 system is essential to its safe adoption for clinical and research applications. Although the conformational dynamics of Cas9 are known to control its ...Controlling the activity of the CRISPR-Cas9 system is essential to its safe adoption for clinical and research applications. Although the conformational dynamics of Cas9 are known to control its enzymatic activity, details of how Cas9 influences the catalytic processes at both nuclease domains remain elusive. Here we report five cryo-electron microscopy structures of the active Cas9 complex along the reaction path at 2.2-2.9 Å resolution. We observed that a large movement in one nuclease domain, triggered by the cognate DNA, results in noticeable changes in the active site of the other domain that is required for metal coordination and catalysis. Furthermore, the conformations synchronize the reaction intermediates, enabling coupled cutting of the two DNA strands. Consistent with the roles of conformations in organizing the active sites, adjustments to the metal-coordination residues lead to altered metal specificity of Cas9 and commonly used Cas9 in cells. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8d2o.cif.gz 8d2o.cif.gz | 271.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8d2o.ent.gz pdb8d2o.ent.gz | 173.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8d2o.json.gz 8d2o.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/d2/8d2o https://data.pdbj.org/pub/pdb/validation_reports/d2/8d2o ftp://data.pdbj.org/pub/pdb/validation_reports/d2/8d2o ftp://data.pdbj.org/pub/pdb/validation_reports/d2/8d2o | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  27144MC  8d2kC  8d2lC  8d2nC  8d2pC  8d2qC C: citing same article ( M: map data used to model this data |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-DNA target strand (5'- ... , 2 types, 2 molecules XT
| #3: DNA chain | Mass: 4021.632 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.) synthetic construct (others) |
|---|---|
| #4: DNA chain | Mass: 3349.197 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.) synthetic construct (others) |
-Protein / RNA chain / DNA chain , 3 types, 3 molecules ABD
| #1: Protein | Mass: 127498.094 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Acidothermus cellulolyticus 11B (bacteria) Acidothermus cellulolyticus 11B (bacteria)Strain: ATCC 43068 / DSM 8971 / 11B / Gene: Acel_1951 / Production host:   Escherichia coli (E. coli) / References: UniProt: A0LWB3 Escherichia coli (E. coli) / References: UniProt: A0LWB3 |
|---|---|
| #2: RNA chain | Mass: 34190.301 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Acidothermus cellulolyticus 11B (bacteria) Acidothermus cellulolyticus 11B (bacteria)Strain: ATCC 43068 / DSM 8971 / 11B / Production host:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| #5: DNA chain | Mass: 3615.395 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.) synthetic construct (others) |
-Non-polymers , 2 types, 8 molecules 


| #6: Chemical | ChemComp-MG / #7: Water | ChemComp-HOH / |  Water Water |
|---|
-Details
| Has ligand of interest | N |
|---|
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method:  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Component |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Source (natural) | Organism:   Acidothermus cellulolyticus 11B (bacteria) Acidothermus cellulolyticus 11B (bacteria) | ||||||||||||||||||||||||
| Source (recombinant) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) | ||||||||||||||||||||||||
| Buffer solution | pH: 7.5 | ||||||||||||||||||||||||
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied : NO / Vitrification applied : NO / Vitrification applied : YES : YES | ||||||||||||||||||||||||
Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source : :  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2200 nm / Nominal defocus min: 1000 nm Bright-field microscopy / Nominal defocus max: 2200 nm / Nominal defocus min: 1000 nm |
| Image recording | Electron dose: 60 e/Å2 / Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) |
| EM imaging optics | Energyfilter name : GIF Bioquantum / Energyfilter slit width: 20 eV : GIF Bioquantum / Energyfilter slit width: 20 eV |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software | Name: RELION / Version: 4 / Category: final Euler assignment | ||||||||||||||||||||||||
CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||
3D reconstruction | Resolution: 2.66 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 169296 / Symmetry type: POINT | ||||||||||||||||||||||||
| Refinement | Cross valid method: NONE Stereochemistry target values: GeoStd + Monomer Library + CDL v1.2 | ||||||||||||||||||||||||
| Displacement parameters | Biso mean: 41.06 Å2 | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller





 PDBj
PDBj
































































