[English] 日本語
 Yorodumi
Yorodumi- PDB-7vos: High-resolution neutron and X-ray joint refined structure of high... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7vos | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | High-resolution neutron and X-ray joint refined structure of high-potential iron-sulfur protein in the oxidized state | ||||||||||||
 Components Components | High-potential iron-sulfur protein | ||||||||||||
 Keywords Keywords | METAL BINDING PROTEIN /  IRON-SULFUR PROTEIN IRON-SULFUR PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationaerobic electron transport chain / 4 iron, 4 sulfur cluster binding /  electron transfer activity / electron transfer activity /  metal ion binding metal ion bindingSimilarity search - Function | ||||||||||||
| Biological species |  Thermochromatium tepidum (bacteria) Thermochromatium tepidum (bacteria) | ||||||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  NEUTRON DIFFRACTION / NEUTRON DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 0.66 Å MOLECULAR REPLACEMENT / Resolution: 0.66 Å | ||||||||||||
 Authors Authors | Hanazono, Y. / Hirano, Y. / Takeda, K. / Kusaka, K. / Tamada, T. / Miki, K. | ||||||||||||
| Funding support |  Japan, 3items Japan, 3items
| ||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: Revisiting the concept of peptide bond planarity in an iron-sulfur protein by neutron structure analysis. Authors: Hanazono, Y. / Hirano, Y. / Takeda, K. / Kusaka, K. / Tamada, T. / Miki, K. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7vos.cif.gz 7vos.cif.gz | 82.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7vos.ent.gz pdb7vos.ent.gz | 63.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7vos.json.gz 7vos.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/vo/7vos https://data.pdbj.org/pub/pdb/validation_reports/vo/7vos ftp://data.pdbj.org/pub/pdb/validation_reports/vo/7vos ftp://data.pdbj.org/pub/pdb/validation_reports/vo/7vos | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5wqqS S: Starting model for refinement |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 8793.851 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Thermochromatium tepidum (bacteria) / References: UniProt: P80176 Thermochromatium tepidum (bacteria) / References: UniProt: P80176 |
|---|
-Non-polymers , 6 types, 145 molecules 


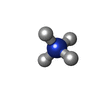







| #2: Chemical | ChemComp-SF4 /  Iron–sulfur cluster Iron–sulfur cluster | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| #3: Chemical | ChemComp-SO4 /  Sulfate Sulfate#4: Chemical | ChemComp-GOL /  Glycerol Glycerol#5: Chemical | ChemComp-NH4 /  Ammonium Ammonium#6: Chemical | ChemComp-D3O / | #7: Chemical | ChemComp-DOD / |  Heavy water Heavy water |
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment |
|
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.83 Å3/Da / Density % sol: 32.62 % |
|---|---|
Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop / pH: 4.5 / Details: 1.8M ammonium sulfate, 0.1M Na citrate buffer |
-Data collection
| Diffraction |
| |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source |
| |||||||||||||||||||||
| Detector |
| |||||||||||||||||||||
| Radiation |
| |||||||||||||||||||||
| Radiation wavelength |
| |||||||||||||||||||||
| Reflection | Biso Wilson estimate: 3.81 Å2 / Entry-ID: 7VOS
| |||||||||||||||||||||
| Reflection shell |
|
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | SU ML: 0.044 / Cross valid method: FREE R-VALUE / Method to determine structure
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 0.66→36.41 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller


 PDBj
PDBj








