[English] 日本語
 Yorodumi
Yorodumi- PDB-7pbw: Cryo-EM structure of light harvesting complex 2 from Rba. sphaeroides. -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7pbw | ||||||
|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of light harvesting complex 2 from Rba. sphaeroides. | ||||||
 Components Components | (Light-harvesting protein B-800/850 ...) x 2 | ||||||
 Keywords Keywords |  PHOTOSYNTHESIS / PHOTOSYNTHESIS /  Purple bacteria / Purple bacteria /  Light harvesting complex 2 / LH2 / Light harvesting complex 2 / LH2 /  Membrane protein / Membrane protein /  Cryo-EM. Cryo-EM. | ||||||
| Function / homology |  Function and homology information Function and homology informationorganelle inner membrane / : / plasma membrane light-harvesting complex /  bacteriochlorophyll binding / bacteriochlorophyll binding /  photosynthesis, light reaction / electron transporter, transferring electrons within the cyclic electron transport pathway of photosynthesis activity / membrane => GO:0016020 / photosynthesis, light reaction / electron transporter, transferring electrons within the cyclic electron transport pathway of photosynthesis activity / membrane => GO:0016020 /  metal ion binding / metal ion binding /  plasma membrane plasma membraneSimilarity search - Function | ||||||
| Biological species |  Cereibacter sphaeroides 2.4.1 (bacteria) Cereibacter sphaeroides 2.4.1 (bacteria) | ||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.1 Å cryo EM / Resolution: 2.1 Å | ||||||
 Authors Authors | Qian, P. / Swainsbury, D.J.K. / Croll, T.I. / Castro-Hartmann, P. / Sader, K. / Divitini, G. / Hunter, C.N. | ||||||
| Funding support |  United Kingdom, 1items United Kingdom, 1items
| ||||||
 Citation Citation |  Journal: Biochemistry / Year: 2021 Journal: Biochemistry / Year: 2021Title: Cryo-EM Structure of the Light-Harvesting 2 Complex at 2.1 Å. Authors: Pu Qian / David J K Swainsbury / Tristan I Croll / Pablo Castro-Hartmann / Giorgio Divitini / Kasim Sader / C Neil Hunter /   Abstract: Light-harvesting 2 (LH2) antenna complexes augment the collection of solar energy in many phototrophic bacteria. Despite its frequent role as a model for such complexes, there has been no three- ...Light-harvesting 2 (LH2) antenna complexes augment the collection of solar energy in many phototrophic bacteria. Despite its frequent role as a model for such complexes, there has been no three-dimensional (3D) structure available for the LH2 from the purple phototroph . We used cryo-electron microscopy (cryo-EM) to determine the 2.1 Å resolution structure of this LH2 antenna, which is a cylindrical assembly of nine αβ heterodimer subunits, each of which binds three bacteriochlorophyll (BChl) molecules and one carotenoid. The high resolution of this structure reveals all of the interpigment and pigment-protein interactions that promote the assembly and energy-transfer properties of this complex. Near the cytoplasmic face of the complex there is a ring of nine BChls, which absorb maximally at 800 nm and are designated as B800; each B800 is coordinated by the N-terminal carboxymethionine of LH2-α, part of a network of interactions with nearby residues on both LH2-α and LH2-β and with the carotenoid. Nine carotenoids, which are spheroidene in the strain we analyzed, snake through the complex, traversing the membrane and interacting with a ring of 18 BChls situated toward the periplasmic side of the complex. Hydrogen bonds with C-terminal aromatic residues modify the absorption of these pigments, which are red-shifted to 850 nm. Overlaps between the macrocycles of the B850 BChls ensure rapid transfer of excitation energy around this ring of pigments, which act as the donors of energy to neighboring LH2 and reaction center light-harvesting 1 (RC-LH1) complexes. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7pbw.cif.gz 7pbw.cif.gz | 282.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7pbw.ent.gz pdb7pbw.ent.gz | Display |  PDB format PDB format | |
| PDBx/mmJSON format |  7pbw.json.gz 7pbw.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/pb/7pbw https://data.pdbj.org/pub/pdb/validation_reports/pb/7pbw ftp://data.pdbj.org/pub/pdb/validation_reports/pb/7pbw ftp://data.pdbj.org/pub/pdb/validation_reports/pb/7pbw | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  13307MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Light-harvesting protein B-800/850 ... , 2 types, 18 molecules AAABACADAEAFAGAHAIBABBBCBDBEBFBGBHBI
| #1: Protein/peptide | Mass: 5275.187 Da / Num. of mol.: 9 / Source method: isolated from a natural source / Source: (natural)  Cereibacter sphaeroides 2.4.1 (bacteria) / References: UniProt: Q3J144 Cereibacter sphaeroides 2.4.1 (bacteria) / References: UniProt: Q3J144#2: Protein/peptide | Mass: 4990.840 Da / Num. of mol.: 9 / Source method: isolated from a natural source / Source: (natural)  Cereibacter sphaeroides 2.4.1 (bacteria) / References: UniProt: Q3J145 Cereibacter sphaeroides 2.4.1 (bacteria) / References: UniProt: Q3J145 |
|---|
-Non-polymers , 5 types, 311 molecules 
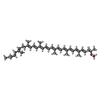







| #3: Chemical | ChemComp-BCL /  Bacteriochlorophyll Bacteriochlorophyll#4: Chemical | ChemComp-7OT / #5: Chemical | ChemComp-LDA /  Lauryldimethylamine oxide Lauryldimethylamine oxide#6: Chemical | ChemComp-CA / #7: Water | ChemComp-HOH / |  Water Water |
|---|
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method:  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Light harvesting complex 2 Light-harvesting complex / Type: COMPLEX / Entity ID: #1-#2 / Source: NATURAL Light-harvesting complex / Type: COMPLEX / Entity ID: #1-#2 / Source: NATURAL |
|---|---|
| Molecular weight | Value: 0.1237 MDa / Experimental value: YES |
| Source (natural) | Organism:  Cereibacter sphaeroides 2.4.1 (bacteria) Cereibacter sphaeroides 2.4.1 (bacteria) |
| Buffer solution | pH: 7.8 Details: The final sample buffer: 20 mM Hepes, pH 7.8, 0.1%LDAO |
| Buffer component | Conc.: 20 mmol / Name: Hepes / Formula: C8H18N2O4S / Formula: C8H18N2O4S |
| Specimen | Conc.: 4.5 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied : NO / Vitrification applied : NO / Vitrification applied : YES : YESDetails: Sample was solubilised and purified with detergent of LDAO |
| Specimen support | Details: easiGlow was used for glow discharge. / Grid material: COPPER / Grid mesh size: 300 divisions/in. / Grid type: Quantifoil R1.2/1.3 |
Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy imaging
Electron microscopy imaging
| Microscopy | Model: FEI TITAN |
|---|---|
| Electron gun | Electron source : :  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD Bright-field microscopy / Nominal magnification: 130000 X / Nominal defocus max: 2200 nm / Nominal defocus min: 800 nm / Cs Bright-field microscopy / Nominal magnification: 130000 X / Nominal defocus max: 2200 nm / Nominal defocus min: 800 nm / Cs : 2.7 mm / Alignment procedure: COMA FREE : 2.7 mm / Alignment procedure: COMA FREE |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Average exposure time: 1.37 sec. / Electron dose: 41.6 e/Å2 / Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Num. of grids imaged: 1 / Num. of real images: 3138 Details: A total doe 41.6 was fractioned into 40 frames, resulting in an electron fluency of 1.04 e/A2/frame. |
| EM imaging optics | Energyfilter name : GIF Bioquantum / Energyfilter slit width: 20 eV : GIF Bioquantum / Energyfilter slit width: 20 eV |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software |
| ||||||||||||||||||||||||||||
CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 1717608 | ||||||||||||||||||||||||||||
| Symmetry | Point symmetry : C9 (9 fold cyclic : C9 (9 fold cyclic ) ) | ||||||||||||||||||||||||||||
3D reconstruction | Resolution: 2.1 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 835641 / Algorithm: BACK PROJECTION / Num. of class averages: 1 / Symmetry type: POINT | ||||||||||||||||||||||||||||
| Refinement | Cross valid method: NONE Stereochemistry target values: GeoStd + Monomer Library + CDL v1.2 | ||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 12.34 Å2 | ||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller










 PDBj
PDBj



