[English] 日本語
 Yorodumi
Yorodumi- PDB-6myt: Avian mitochondrial complex II with Atpenin A5 bound, sidechain i... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6myt | ||||||
|---|---|---|---|---|---|---|---|
| Title | Avian mitochondrial complex II with Atpenin A5 bound, sidechain in pocket | ||||||
 Components Components |
| ||||||
 Keywords Keywords | OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR /  Complex II / Complex II /  membrane protein / membrane protein /  heme protein / OXIDOREDUCTASE-OXIDOREDUCTASE INHIBITOR complex heme protein / OXIDOREDUCTASE-OXIDOREDUCTASE INHIBITOR complex | ||||||
| Function / homology |  Function and homology information Function and homology informationCitric acid cycle (TCA cycle) / The tricarboxylic acid cycle /  succinate dehydrogenase complex / succinate dehydrogenase complex /  succinate dehydrogenase activity / succinate metabolic process / mitochondrial electron transport, succinate to ubiquinone / mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone) / succinate dehydrogenase (quinone) activity / : / succinate dehydrogenase activity / succinate metabolic process / mitochondrial electron transport, succinate to ubiquinone / mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone) / succinate dehydrogenase (quinone) activity / : /  succinate dehydrogenase ...Citric acid cycle (TCA cycle) / The tricarboxylic acid cycle / succinate dehydrogenase ...Citric acid cycle (TCA cycle) / The tricarboxylic acid cycle /  succinate dehydrogenase complex / succinate dehydrogenase complex /  succinate dehydrogenase activity / succinate metabolic process / mitochondrial electron transport, succinate to ubiquinone / mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone) / succinate dehydrogenase (quinone) activity / : / succinate dehydrogenase activity / succinate metabolic process / mitochondrial electron transport, succinate to ubiquinone / mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone) / succinate dehydrogenase (quinone) activity / : /  succinate dehydrogenase / oxidoreductase activity, acting on the CH-CH group of donors / succinate dehydrogenase / oxidoreductase activity, acting on the CH-CH group of donors /  ubiquinone binding / 3 iron, 4 sulfur cluster binding / ubiquinone binding / 3 iron, 4 sulfur cluster binding /  aerobic respiration / aerobic respiration /  tricarboxylic acid cycle / respiratory electron transport chain / tricarboxylic acid cycle / respiratory electron transport chain /  mitochondrial membrane / 2 iron, 2 sulfur cluster binding / mitochondrial membrane / 2 iron, 2 sulfur cluster binding /  flavin adenine dinucleotide binding / 4 iron, 4 sulfur cluster binding / flavin adenine dinucleotide binding / 4 iron, 4 sulfur cluster binding /  mitochondrial inner membrane / mitochondrial inner membrane /  electron transfer activity / electron transfer activity /  heme binding / heme binding /  metal ion binding metal ion bindingSimilarity search - Function | ||||||
| Biological species |   Gallus gallus (chicken) Gallus gallus (chicken) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.27 Å MOLECULAR REPLACEMENT / Resolution: 2.27 Å | ||||||
 Authors Authors | Berry, E.A. / Huang, L.-S. | ||||||
| Funding support |  United States, 1items United States, 1items
| ||||||
 Citation Citation |  Journal: Biochim Biophys Acta Proteins Proteom / Year: 2021 Journal: Biochim Biophys Acta Proteins Proteom / Year: 2021Title: Crystallographic investigation of the ubiquinone binding site of respiratory Complex II and its inhibitors. Authors: Huang, L.S. / Lummen, P. / Berry, E.A. #1:  Journal: J. Biol. Chem. / Year: 2006 Journal: J. Biol. Chem. / Year: 2006Title: 3-nitropropionic acid is a suicide inhibitor of mitochondrial respiration that, upon oxidation by complex II, forms a covalent adduct with a catalytic base arginine in the active site of the enzyme. Authors: Huang, L.S. / Sun, G. / Cobessi, D. / Wang, A.C. / Shen, J.T. / Tung, E.Y. / Anderson, V.E. / Berry, E.A. #2:  Journal: Biochim. Biophys. Acta / Year: 2006 Journal: Biochim. Biophys. Acta / Year: 2006Title: Crystallographic studies of the binding of ligands to the dicarboxylate site of Complex II, and the identity of the ligand in the "oxaloacetate-inhibited" state. Authors: Huang, L.S. / Shen, J.T. / Wang, A.C. / Berry, E.A. #3: Journal: Acta Crystallogr. D Biol. Crystallogr. / Year: 2005 Title: Crystallization of mitochondrial respiratory complex II from chicken heart: a membrane-protein complex diffracting to 2.0 A. Authors: Huang, L.S. / Borders, T.M. / Shen, J.T. / Wang, C.J. / Berry, E.A. #4:  Journal: Cell / Year: 2005 Journal: Cell / Year: 2005Title: Crystal structure of mitochondrial respiratory membrane protein complex II. Authors: Sun, F. / Huo, X. / Zhai, Y. / Wang, A. / Xu, J. / Su, D. / Bartlam, M. / Rao, Z. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6myt.cif.gz 6myt.cif.gz | 480 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6myt.ent.gz pdb6myt.ent.gz | 386.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6myt.json.gz 6myt.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/my/6myt https://data.pdbj.org/pub/pdb/validation_reports/my/6myt ftp://data.pdbj.org/pub/pdb/validation_reports/my/6myt ftp://data.pdbj.org/pub/pdb/validation_reports/my/6myt | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6myoC  6mypC  6myqC  6myrC  6mysC  6myuC  1yq3S S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Succinate dehydrogenase [ubiquinone] ... , 3 types, 3 molecules ABD
| #1: Protein | Mass: 68256.922 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Gallus gallus (chicken) / Organ: heart Gallus gallus (chicken) / Organ: heart References: UniProt: F1NPJ4, UniProt: Q9YHT1*PLUS,  succinate dehydrogenase succinate dehydrogenase |
|---|---|
| #2: Protein | Mass: 28685.221 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Gallus gallus (chicken) / Organ: heart Gallus gallus (chicken) / Organ: heart / References: UniProt: Q9YHT2, / References: UniProt: Q9YHT2,  succinate dehydrogenase succinate dehydrogenase |
| #4: Protein | Mass: 10971.604 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Gallus gallus (chicken) / Organ: heart Gallus gallus (chicken) / Organ: heart / References: UniProt: Q5ZIS0 / References: UniProt: Q5ZIS0 |
-Protein , 1 types, 1 molecules C
| #3: Protein | Mass: 15391.153 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Gallus gallus (chicken) / Organ: heart Gallus gallus (chicken) / Organ: heart References: UniProt: D0VWW3, UniProt: A0A3Q2U2Y6*PLUS,  succinate dehydrogenase succinate dehydrogenase |
|---|
-Non-polymers , 13 types, 270 molecules 







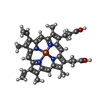














| #5: Chemical | ChemComp-FAD /  Flavin adenine dinucleotide Flavin adenine dinucleotide | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| #6: Chemical | ChemComp-OAA /  Oxaloacetic acid Oxaloacetic acid | ||||||||||||||||||
| #7: Chemical | ChemComp-K / | ||||||||||||||||||
| #8: Chemical | ChemComp-UNL / Num. of mol.: 59 / Source method: obtained synthetically #9: Chemical | ChemComp-PEG / |  Diethylene glycol Diethylene glycol#10: Chemical | ChemComp-FES / |  Iron–sulfur cluster Iron–sulfur cluster#11: Chemical | ChemComp-SF4 / |  Iron–sulfur cluster Iron–sulfur cluster#12: Chemical | ChemComp-F3S / |  Iron–sulfur cluster Iron–sulfur cluster#13: Chemical | ChemComp-AT5 / | #14: Chemical | ChemComp-HEM / |  Heme B Heme B#15: Chemical | ChemComp-UMQ / | #16: Chemical | ChemComp-3PE / |  Phosphatidylethanolamine Phosphatidylethanolamine#17: Water | ChemComp-HOH / |  Water Water |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.46 Å3/Da / Density % sol: 64.43 % |
|---|---|
Crystal grow | Temperature: 277 K / Method: vapor diffusion, sitting drop / pH: 7.5 Details: 47 g/L PEG3350, 30 mL/L PEG400, 23 mL/L isopropanol, 0.05 M HEPES sodium, 0.01 M Tris-HCl, 3.1 mM manganese chloride, 0.62 mM magnesium chloride, 1.5 mM sodium azide, 0.25 mM sodium EDTA, 10 ...Details: 47 g/L PEG3350, 30 mL/L PEG400, 23 mL/L isopropanol, 0.05 M HEPES sodium, 0.01 M Tris-HCl, 3.1 mM manganese chloride, 0.62 mM magnesium chloride, 1.5 mM sodium azide, 0.25 mM sodium EDTA, 10 g/L octyl beta-D-glucoside, undecyl-beta-D-maltoside, crystal was soaked with Atpenin A5 after crystallization |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ALS ALS  / Beamline: 5.0.2 / Wavelength: 1 Å / Beamline: 5.0.2 / Wavelength: 1 Å |
| Detector | Type: ADSC QUANTUM 315r / Detector: CCD / Date: Apr 1, 2007 |
| Radiation | Monochromator: liquid nitrogen-cooled dual crystal Si(111) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 1 Å / Relative weight: 1 : 1 Å / Relative weight: 1 |
| Reflection | Resolution: 2.27→25.23 Å / Num. obs: 70243 / % possible obs: 87.6 % / Redundancy: 2.15 % / Biso Wilson estimate: 42.29 Å2 / Rmerge(I) obs: 0.145 / Net I/σ(I): 4.1 |
| Reflection shell | Resolution: 2.27→2.34 Å / Redundancy: 1.53 % / Rmerge(I) obs: 0.557 / Mean I/σ(I) obs: 1.5 / % possible all: 67.8 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1YQ3 Resolution: 2.27→25.19 Å / SU ML: 0.39 / Cross valid method: FREE R-VALUE / σ(F): 1.33 / Phase error: 33.36 / Stereochemistry target values: ML
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 1 Å / VDW probe radii: 1.2 Å / Solvent model: FLAT BULK SOLVENT MODEL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 60.5814 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.27→25.19 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Origin x: 20.4176 Å / Origin y: 18.5239 Å / Origin z: 110.1542 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group | Selection details: ALL |
 Movie
Movie Controller
Controller





















 PDBj
PDBj
























