[English] 日本語
 Yorodumi
Yorodumi- PDB-4v8k: Crystal structure of the LH1-RC complex from Thermochromatium tep... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4v8k | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crystal structure of the LH1-RC complex from Thermochromatium tepidum in P21 form | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords |  PHOTOSYNTHESIS PHOTOSYNTHESIS | |||||||||
| Function / homology |  Function and homology information Function and homology informationorganelle inner membrane / plasma membrane-derived chromatophore membrane / plasma membrane light-harvesting complex /  bacteriochlorophyll binding / photosynthetic electron transport in photosystem II / bacteriochlorophyll binding / photosynthetic electron transport in photosystem II /  photosynthesis, light reaction / electron transporter, transferring electrons within the cyclic electron transport pathway of photosynthesis activity / photosynthesis, light reaction / electron transporter, transferring electrons within the cyclic electron transport pathway of photosynthesis activity /  photosynthesis / photosynthesis /  electron transfer activity / iron ion binding ...organelle inner membrane / plasma membrane-derived chromatophore membrane / plasma membrane light-harvesting complex / electron transfer activity / iron ion binding ...organelle inner membrane / plasma membrane-derived chromatophore membrane / plasma membrane light-harvesting complex /  bacteriochlorophyll binding / photosynthetic electron transport in photosystem II / bacteriochlorophyll binding / photosynthetic electron transport in photosystem II /  photosynthesis, light reaction / electron transporter, transferring electrons within the cyclic electron transport pathway of photosynthesis activity / photosynthesis, light reaction / electron transporter, transferring electrons within the cyclic electron transport pathway of photosynthesis activity /  photosynthesis / photosynthesis /  electron transfer activity / iron ion binding / electron transfer activity / iron ion binding /  heme binding / heme binding /  membrane / membrane /  metal ion binding / metal ion binding /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |  Thermochromatium tepidum (bacteria) Thermochromatium tepidum (bacteria) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MIR / Resolution: 3.006 Å MIR / Resolution: 3.006 Å | |||||||||
 Authors Authors | Niwa, S. / Takeda, K. / Wang-Otomo, Z.-Y. / Miki, K. | |||||||||
 Citation Citation |  Journal: Nature / Year: 2014 Journal: Nature / Year: 2014Title: Structure of the LH1-RC complex from Thermochromatium tepidum at 3.0 angstrom Authors: Niwa, S. / Yu, L.J. / Takeda, K. / Hirano, Y. / Kawakami, T. / Wang-Otomo, Z.Y. / Miki, K. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4v8k.cif.gz 4v8k.cif.gz | 1.9 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4v8k.ent.gz pdb4v8k.ent.gz | Display |  PDB format PDB format | |
| PDBx/mmJSON format |  4v8k.json.gz 4v8k.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/v8/4v8k https://data.pdbj.org/pub/pdb/validation_reports/v8/4v8k ftp://data.pdbj.org/pub/pdb/validation_reports/v8/4v8k ftp://data.pdbj.org/pub/pdb/validation_reports/v8/4v8k | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
-Photosynthetic reaction center ... , 4 types, 8 molecules ACBCALBLAMBMAHBH
| #1: Protein | Mass: 43177.531 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Thermochromatium tepidum (bacteria) / References: UniProt: D2Z0P5 Thermochromatium tepidum (bacteria) / References: UniProt: D2Z0P5#2: Protein | Mass: 31520.771 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Thermochromatium tepidum (bacteria) / References: UniProt: D2Z0P3 Thermochromatium tepidum (bacteria) / References: UniProt: D2Z0P3#3: Protein | Mass: 36605.715 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Thermochromatium tepidum (bacteria) / References: UniProt: A8ASG6 Thermochromatium tepidum (bacteria) / References: UniProt: A8ASG6#4: Protein | Mass: 28213.305 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Thermochromatium tepidum (bacteria) / References: UniProt: D2Z0P9 Thermochromatium tepidum (bacteria) / References: UniProt: D2Z0P9 |
|---|
-Protein / Protein/peptide , 2 types, 64 molecules AAADAFAIAKAOAQASAUAWAYA1A3A5A7A9BABDBFBIBKBOBQBSBUBWBYB1B3B5...
| #5: Protein | Mass: 7034.442 Da / Num. of mol.: 32 / Source method: isolated from a natural source / Source: (natural)  Thermochromatium tepidum (bacteria) / References: UniProt: D2Z0P2 Thermochromatium tepidum (bacteria) / References: UniProt: D2Z0P2#6: Protein/peptide | Mass: 5534.452 Da / Num. of mol.: 32 / Source method: isolated from a natural source / Source: (natural)  Thermochromatium tepidum (bacteria) / References: UniProt: D2Z0P1 Thermochromatium tepidum (bacteria) / References: UniProt: D2Z0P1 |
|---|
-Non-polymers , 11 types, 190 molecules 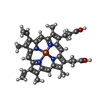


















| #7: Chemical | ChemComp-HEM /  Heme B Heme B#8: Chemical | ChemComp-CA / #9: Chemical | ChemComp-BCL /  Bacteriochlorophyll Bacteriochlorophyll#10: Chemical | ChemComp-BPH /  Pheophytin Pheophytin#11: Chemical | #12: Chemical |  Iron Iron#13: Chemical |  Vitamin K2 Vitamin K2#14: Chemical | ChemComp-CRT / #15: Chemical | ChemComp-PEF /  Phosphatidylethanolamine Phosphatidylethanolamine#16: Chemical | ChemComp-PO4 /  Phosphate Phosphate#17: Water | ChemComp-HOH / |  Water Water |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.56 Å3/Da / Density % sol: 65.47 % |
|---|---|
Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop / pH: 7.4 Details: 20mM Tris-HCl (pH 7.4), 50mM CaCl2, 0.54% DPC, 2% glycerol, 14% PEG 3000 , VAPOR DIFFUSION, SITTING DROP, temperature 293K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SPring-8 SPring-8  / Beamline: BL44XU / Wavelength: 0.9 Å / Beamline: BL44XU / Wavelength: 0.9 Å |
| Detector | Type: RAYONIX MX225HE / Detector: CCD / Date: Jan 30, 2011 |
| Radiation | Monochromator: Si111 / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 0.9 Å / Relative weight: 1 : 0.9 Å / Relative weight: 1 |
| Reflection | Resolution: 3→50 Å / Num. all: 167629 / Num. obs: 167629 / % possible obs: 88.5 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 5.4 % / Biso Wilson estimate: 55.76 Å2 / Rsym value: 0.087 / Net I/σ(I): 9.14 |
| Reflection shell | Resolution: 3→3.11 Å / Redundancy: 2.1 % / Mean I/σ(I) obs: 1.73 / Rsym value: 0.377 / % possible all: 71.8 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MIR / Resolution: 3.006→43.794 Å / FOM work R set: 0.5941 / SU ML: 0.8 / σ(F): 3 / Phase error: 45.07 / Stereochemistry target values: MLHL MIR / Resolution: 3.006→43.794 Å / FOM work R set: 0.5941 / SU ML: 0.8 / σ(F): 3 / Phase error: 45.07 / Stereochemistry target values: MLHL
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 134.85 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 3.006→43.794 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Total num. of bins used: 10
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj











