+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
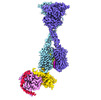
| Title | Consensus map of human CaSR-miniGis complex in nanodiscs | |||||||||
 Map data Map data | Consensus map of human CaSR-miniGis complex in nanodiscs | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Calcium-sensing receptor / Calcium-sensing receptor /  G-protein-coupled receptor / G-protein-coupled receptor /  G protein / G protein /  signal transduction / signal transduction /  MEMBRANE PROTEIN MEMBRANE PROTEIN | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.6 Å cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Zuo H / Park J / Frangaj A / Ye J / Lu G / Manning JJ / Asher WB / Lu Z / Hu G / Wang L ...Zuo H / Park J / Frangaj A / Ye J / Lu G / Manning JJ / Asher WB / Lu Z / Hu G / Wang L / Mendez J / Eng E / Zhang Z / Lin X / Grasucci R / Hendrickson WA / Clarke OB / Javitch JA / Conigrave AD / Fan QR | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Promiscuous G-protein activation by the calcium-sensing receptor. Authors: Hao Zuo / Jinseo Park / Aurel Frangaj / Jianxiang Ye / Guanqi Lu / Jamie J Manning / Wesley B Asher / Zhengyuan Lu / Guo-Bin Hu / Liguo Wang / Joshua Mendez / Edward Eng / Zhening Zhang / ...Authors: Hao Zuo / Jinseo Park / Aurel Frangaj / Jianxiang Ye / Guanqi Lu / Jamie J Manning / Wesley B Asher / Zhengyuan Lu / Guo-Bin Hu / Liguo Wang / Joshua Mendez / Edward Eng / Zhening Zhang / Xin Lin / Robert Grassucci / Wayne A Hendrickson / Oliver B Clarke / Jonathan A Javitch / Arthur D Conigrave / Qing R Fan /   Abstract: The human calcium-sensing receptor (CaSR) detects fluctuations in the extracellular Ca concentration and maintains Ca homeostasis. It also mediates diverse cellular processes not associated with Ca ...The human calcium-sensing receptor (CaSR) detects fluctuations in the extracellular Ca concentration and maintains Ca homeostasis. It also mediates diverse cellular processes not associated with Ca balance. The functional pleiotropy of CaSR arises in part from its ability to signal through several G-protein subtypes. We determined structures of CaSR in complex with G proteins from three different subfamilies: G, G and G. We found that the homodimeric CaSR of each complex couples to a single G protein through a common mode. This involves the C-terminal helix of each Gα subunit binding to a shallow pocket that is formed in one CaSR subunit by all three intracellular loops (ICL1-ICL3), an extended transmembrane helix 3 and an ordered C-terminal region. G-protein binding expands the transmembrane dimer interface, which is further stabilized by phospholipid. The restraint imposed by the receptor dimer, in combination with ICL2, enables G-protein activation by facilitating conformational transition of Gα. We identified a single Gα residue that determines G and G versus G selectivity. The length and flexibility of ICL2 allows CaSR to bind all three Gα subtypes, thereby conferring capacity for promiscuous G-protein coupling. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43897.map.gz emd_43897.map.gz | 255.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43897-v30.xml emd-43897-v30.xml emd-43897.xml emd-43897.xml | 21.6 KB 21.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_43897_fsc.xml emd_43897_fsc.xml | 17 KB | Display |  FSC data file FSC data file |
| Images |  emd_43897.png emd_43897.png | 76 KB | ||
| Filedesc metadata |  emd-43897.cif.gz emd-43897.cif.gz | 5.3 KB | ||
| Others |  emd_43897_half_map_1.map.gz emd_43897_half_map_1.map.gz emd_43897_half_map_2.map.gz emd_43897_half_map_2.map.gz | 475.4 MB 475.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43897 http://ftp.pdbj.org/pub/emdb/structures/EMD-43897 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43897 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43897 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_43897.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43897.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Consensus map of human CaSR-miniGis complex in nanodiscs | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.825 Å | ||||||||||||||||||||||||||||||||||||
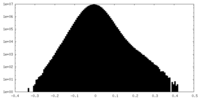
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map A
| File | emd_43897_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
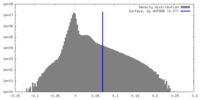
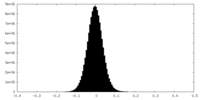
| Density Histograms |
-Half map: Half map B
| File | emd_43897_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human CaSR in complex with chimeric Gs (miniGis) protein
| Entire | Name: Human CaSR in complex with chimeric Gs (miniGis) protein |
|---|---|
| Components |
|
-Supramolecule #1: Human CaSR in complex with chimeric Gs (miniGis) protein
| Supramolecule | Name: Human CaSR in complex with chimeric Gs (miniGis) protein type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 276 KDa |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.9 mg/mL | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||||||||
| Grid | Model: Quantifoil R0.6/1 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Support film - Film thickness: 50 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 25 sec. / Pretreatment - Atmosphere: OTHER | |||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: The sample was blotted for 6s before plunge-frozen.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 1.9000000000000001 µm / Nominal defocus min: 1.1 µm / Nominal magnification: 105000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 1.9000000000000001 µm / Nominal defocus min: 1.1 µm / Nominal magnification: 105000 |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 15 eV |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Temperature | Max: 100.0 K |
| Software | Name: EPU |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 11520 pixel / Digitization - Dimensions - Height: 8184 pixel / Number grids imaged: 1 / Number real images: 29091 / Average exposure time: 2.72 sec. / Average electron dose: 67.9 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Software | Name:  Coot (ver. 0.9.8.1) Coot (ver. 0.9.8.1) | ||||||||||||
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
 Movie
Movie Controller
Controller































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)