[English] 日本語
 Yorodumi
Yorodumi- EMDB-42637: Human LINE-1 retrotransposon ORF2 protein engaged with template R... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 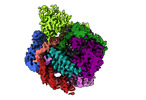 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human LINE-1 retrotransposon ORF2 protein engaged with template RNA in elongation state | |||||||||
 Map data Map data | Human LINE-1 retrotransposon ORF2 protein engaged with template RNA in elongation state | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  LINE-1 / LINE-1 /  retrotransposon / retrotransposon /  reverse transcriptase / retroelement / reverse transcriptase / retroelement /  RNA BINDING PROTEIN / RNA BINDING PROTEIN /  RNA / RNA BINDING PROTEIN-RNA-DNA complex RNA / RNA BINDING PROTEIN-RNA-DNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology information retrotransposition / nucleic acid metabolic process / retrotransposition / nucleic acid metabolic process /  type II site-specific deoxyribonuclease activity / type II site-specific deoxyribonuclease activity /  Hydrolases; Acting on ester bonds; Endodeoxyribonucleases producing 5'-phosphomonoesters / Hydrolases; Acting on ester bonds; Endodeoxyribonucleases producing 5'-phosphomonoesters /  RNA-directed DNA polymerase / RNA-directed DNA polymerase /  RNA-directed DNA polymerase activity / DNA recombination / RNA-directed DNA polymerase activity / DNA recombination /  RNA binding / RNA binding /  metal ion binding metal ion bindingSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.2 Å cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Thawani A / Florez Ariza AJ / Collins K / Nogales E | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Template and target-site recognition by human LINE-1 in retrotransposition. Authors: Akanksha Thawani / Alfredo Jose Florez Ariza / Eva Nogales / Kathleen Collins /  Abstract: The long interspersed element-1 (LINE-1, hereafter L1) retrotransposon has generated nearly one-third of the human genome and serves as an active source of genetic diversity and human disease. L1 ...The long interspersed element-1 (LINE-1, hereafter L1) retrotransposon has generated nearly one-third of the human genome and serves as an active source of genetic diversity and human disease. L1 spreads through a mechanism termed target-primed reverse transcription, in which the encoded enzyme (ORF2p) nicks the target DNA to prime reverse transcription of its own or non-self RNAs. Here we purified full-length L1 ORF2p and biochemically reconstituted robust target-primed reverse transcription with template RNA and target-site DNA. We report cryo-electron microscopy structures of the complete human L1 ORF2p bound to structured template RNAs and initiating cDNA synthesis. The template polyadenosine tract is recognized in a sequence-specific manner by five distinct domains. Among them, an RNA-binding domain bends the template backbone to allow engagement of an RNA hairpin stem with the L1 ORF2p C-terminal segment. Moreover, structure and biochemical reconstitutions demonstrate an unexpected target-site requirement: L1 ORF2p relies on upstream single-stranded DNA to position the adjacent duplex in the endonuclease active site for nicking of the longer DNA strand, with a single nick generating a staggered DNA break. Our research provides insights into the mechanism of ongoing transposition in the human genome and informs the engineering of retrotransposon proteins for gene therapy. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42637.map.gz emd_42637.map.gz | 5.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42637-v30.xml emd-42637-v30.xml emd-42637.xml emd-42637.xml | 18 KB 18 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42637_fsc.xml emd_42637_fsc.xml | 8.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_42637.png emd_42637.png | 81.7 KB | ||
| Filedesc metadata |  emd-42637.cif.gz emd-42637.cif.gz | 6.9 KB | ||
| Others |  emd_42637_half_map_1.map.gz emd_42637_half_map_1.map.gz emd_42637_half_map_2.map.gz emd_42637_half_map_2.map.gz | 40.8 MB 40.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42637 http://ftp.pdbj.org/pub/emdb/structures/EMD-42637 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42637 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42637 | HTTPS FTP |
-Related structure data
| Related structure data |  8uw3MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42637.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42637.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human LINE-1 retrotransposon ORF2 protein engaged with template RNA in elongation state | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.81 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half Map 2
| File | emd_42637_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map 1
| File | emd_42637_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human LINE-1 retrotransposon ORF2 protein in elongation stage
| Entire | Name: Human LINE-1 retrotransposon ORF2 protein in elongation stage |
|---|---|
| Components |
|
-Supramolecule #1: Human LINE-1 retrotransposon ORF2 protein in elongation stage
| Supramolecule | Name: Human LINE-1 retrotransposon ORF2 protein in elongation stage type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 175 KDa |
-Macromolecule #1: LINE-1 retrotransposable element ORF2 protein
| Macromolecule | Name: LINE-1 retrotransposable element ORF2 protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 149.300391 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MTGSNSHITI LTLNINGLNS AIKRHRLASW IKSQDPSVCC IQETHLTCRD THRLKIKGWR KIYQANGKQK KAGVAILVSD KTDFKPTKI KRDKEGHYIM VKGSIQQEEL TILNIYAPNT GAPRFIKQVL SDLQRDLDSH TLIMGDFNTP LSTLDRSTRQ K VNKDTQEL ...String: MTGSNSHITI LTLNINGLNS AIKRHRLASW IKSQDPSVCC IQETHLTCRD THRLKIKGWR KIYQANGKQK KAGVAILVSD KTDFKPTKI KRDKEGHYIM VKGSIQQEEL TILNIYAPNT GAPRFIKQVL SDLQRDLDSH TLIMGDFNTP LSTLDRSTRQ K VNKDTQEL NSALHQADLI DIYRTLHPKS TEYTFFSAPH HTYSKIDHIV GSKALLSKCK RTEIITNYLS DHSAIKLELR IK NLTQSRS TTWKLNNLLL NDYWVHNEMK AEIKMFFETN ENKDTTYQNL WDAFKAVCRG KFIALNAHKR KQERSKIDTL TSQ LKELEK QEQTHSKASR RQEITKIRAE LKEIETQKTL QKINESRSWF FERINKIDRP LARLIKKKRE KNQIDTIKND KGDI TTDPT EIQTTIREYY KHLYANKLEN LEEMDKFLDT YTLPRLNQEE VESLNRPITG SEIVAIINSL PTKKSPGPDG FTAEF YQRY KEELVPFLLK LFQSIEKEGI LPNSFYEASI ILIPKPGRDT TKKENFRPIS LMNIDAKILN KILANRIQQH IKKLIH HDQ VGFIPGMQGW FNIRKSINVI QHINRAKDKN HMIISIDAEK AFDKIQQPFM LKTLNKLGID GTYFKIIRAI YDKPTAN II LNGQKLEAFP LKTGTRQGCP LSPLLFNIVL EVLARAIRQE KEIKGIQLGK EEVKLSLFAD DMIVYLENPI VSAQNLLK L ISNFSKVSGY KINVQKSQAF LYTNNRQTES QIMGELPFTI ASKRIKYLGI QLTRDVKDLF KENYKPLLKE IKEETNKWK NIPCSWVGRI NIVKMAILPK VIYRFNAIPI KLPMTFFTEL EKTTLKFIWN QKRARIAKSI LSQKNKAGGI TLPDFKLYYK ATVTKTAWY WYQNRDIDQW NRTEPSEIMP HIYNYLIFDK PEKNKQWGKD SLFNKWCWEN WLAICRKLKL DPFLTPYTKI N SRWIKDLN VKPKTIKTLE ENLGITIQDI GVGKDFMSKT PKAMATKDKI DKWDLIKLKS FCTAKETTIR VNRQPTTWEK IF ATYSSDK GLISRIYNEL KQIYKKKTNN PIKKWAKDMN RHFSKEDIYA AKKHMKKCSS SLAIREMQIK TTMRYHLTPV RMA IIKKSG NNRCWRGCGE IGTLLHCWWD CKLVQPLWKS VWRFLRDLEL EIPFDPAIPL LGIYPNEYKS CCYKDTCTRM FIAA LFTIA KTWNQPKCPT MIDWIKKMWH IYTMEYYAAI KNDEFISFVG TWMKLETIIL SKLSQEQKTK HRIFSLIGGN UniProtKB: LINE-1 retrotransposable element ORF2 protein |
-Macromolecule #2: Template RNA
| Macromolecule | Name: Template RNA / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 23.956451 KDa |
| Sequence | String: GCAGACUGCG UAGGCGUGCA GGAACCUGCA CGCCUACGCA GUCUGCAAAA AAAAAAAAAA ACAAUAUCGG CGCG |
-Macromolecule #3: Complementary DNA
| Macromolecule | Name: Complementary DNA / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 3.951586 KDa |
| Sequence | String: (DC)(DG)(DC)(DG)(DC)(DC)(DG)(DA)(DT)(DA) (DT)(DT)(DDG) |
-Macromolecule #4: THYMIDINE-5'-TRIPHOSPHATE
| Macromolecule | Name: THYMIDINE-5'-TRIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 1 / Formula: TTP |
|---|---|
| Molecular weight | Theoretical: 482.168 Da |
| Chemical component information |  ChemComp-TTP: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 105000 Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 11520 pixel / Digitization - Dimensions - Height: 8184 pixel / Number grids imaged: 2 / Number real images: 23878 / Average exposure time: 1.0 sec. / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z
Z Y
Y X
X


















