+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of the Helicobacter pylori CagYdAP OMC | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords |  T4SS / T4SS /  protein translocation / protein translocation /  MEMBRANE PROTEIN MEMBRANE PROTEIN | ||||||||||||
| Biological species |   Helicobacter pylori 26695 (bacteria) Helicobacter pylori 26695 (bacteria) | ||||||||||||
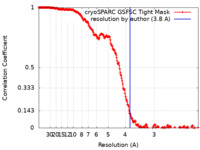
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.8 Å cryo EM / Resolution: 3.8 Å | ||||||||||||
 Authors Authors | Roberts JR | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Life Sci Alliance / Year: 2024 Journal: Life Sci Alliance / Year: 2024Title: Subdomains of the Cag T4SS outer membrane core complex exhibit structural independence. Authors: Jacquelyn R Roberts / Sirena C Tran / Arwen E Frick-Cheng / Kaeli N Bryant / Chiamaka D Okoye / W Hayes McDonald / Timothy L Cover / Melanie D Ohi /  Abstract: The Cag type IV secretion system (Cag T4SS) has an important role in the pathogenesis of gastric cancer. The Cag T4SS outer membrane core complex (OMCC) is organized into three regions: a 14-fold ...The Cag type IV secretion system (Cag T4SS) has an important role in the pathogenesis of gastric cancer. The Cag T4SS outer membrane core complex (OMCC) is organized into three regions: a 14-fold symmetric outer membrane cap (OMC) composed of CagY, CagX, CagT, CagM, and Cag3; a 17-fold symmetric periplasmic ring (PR) composed of CagY and CagX; and a stalk with unknown composition. We investigated how CagT, CagM, and a conserved antenna projection (AP) region of CagY contribute to the structural organization of the OMCC. Single-particle cryo-EM analyses showed that complexes purified from Δ or Δ mutants no longer had organized OMCs, but the PRs remained structured. OMCCs purified from a CagY antenna projection mutant (CagYAP) were structurally similar to WT OMCCs, except for the absence of the α-helical antenna projection. These results indicate that CagY and CagX are sufficient for maintaining a stable PR, but the organization of the OMC requires CagY, CagX, CagM, and CagT. Our results highlight an unexpected structural independence of two major subdomains of the Cag T4SS OMCC. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42290.map.gz emd_42290.map.gz | 979.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42290-v30.xml emd-42290-v30.xml emd-42290.xml emd-42290.xml | 18.6 KB 18.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42290_fsc.xml emd_42290_fsc.xml | 21.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_42290.png emd_42290.png | 191.4 KB | ||
| Filedesc metadata |  emd-42290.cif.gz emd-42290.cif.gz | 5.8 KB | ||
| Others |  emd_42290_half_map_1.map.gz emd_42290_half_map_1.map.gz emd_42290_half_map_2.map.gz emd_42290_half_map_2.map.gz | 962.8 MB 962.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42290 http://ftp.pdbj.org/pub/emdb/structures/EMD-42290 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42290 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42290 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_42290.map.gz / Format: CCP4 / Size: 1 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42290.map.gz / Format: CCP4 / Size: 1 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_42290_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_42290_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Helicobacter pylori CagYdAP OMC
| Entire | Name: Helicobacter pylori CagYdAP OMC |
|---|---|
| Components |
|
-Supramolecule #1: Helicobacter pylori CagYdAP OMC
| Supramolecule | Name: Helicobacter pylori CagYdAP OMC / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Helicobacter pylori 26695 (bacteria) / Strain: 26695 / Location in cell: membrane Helicobacter pylori 26695 (bacteria) / Strain: 26695 / Location in cell: membrane |
-Macromolecule #1: Type IV secretion system apparatus protein Cag3
| Macromolecule | Name: Type IV secretion system apparatus protein Cag3 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Helicobacter pylori 26695 (bacteria) / Strain: 26695 Helicobacter pylori 26695 (bacteria) / Strain: 26695 |
| Sequence | String: MFRKLATAVS LIGLLTSNTL YAKEISEADK VIKATKETKE TKKEAKRLKK EAKQRQQIPD HKKPQYVSV DDTKTQALFD IYDTLNVNDK SFGDWFGNSA LKDKTYLYAM DLLDYNNYLS I ENPIIKTR AMGTYADLII ITGSLEQVNG YYNILKALNK RNAKFVLKIN ...String: MFRKLATAVS LIGLLTSNTL YAKEISEADK VIKATKETKE TKKEAKRLKK EAKQRQQIPD HKKPQYVSV DDTKTQALFD IYDTLNVNDK SFGDWFGNSA LKDKTYLYAM DLLDYNNYLS I ENPIIKTR AMGTYADLII ITGSLEQVNG YYNILKALNK RNAKFVLKIN ENMPYAQATF LR VPKRSDP NAHTLDKGAS IDENKLFEQQ KKMYFNYAND VICRPDDEVC SPLRDEMVAM PTS DSVTQK PNIIAPYSLY RLKETNNANE AQPSPYATQT APENSKEKLI EELIANSQLV ANEE EREKK LLAEKEKQEA ELAKYKLKDL ENQKKLKALE AELKKKNAKK PRVVEVPVSP QTSNS DETM RVVKEKENYN GLLVDKETTI KRSYEGTLIS ENSYSKKTPL NPNDLRSLEE EIKSYY IKS NGLCYTNGIN LYVKIKNDPY KEGMLCGYES VQNLLSPLKD KLKYDKQKLQ KALLKDS K |
-Macromolecule #2: Type IV secretion system apparatus protein CagM
| Macromolecule | Name: Type IV secretion system apparatus protein CagM / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Helicobacter pylori 26695 (bacteria) / Strain: 26695 Helicobacter pylori 26695 (bacteria) / Strain: 26695 |
| Sequence | String: MLAKIVFSSL VAFGVLSAN V EQFGSFFN EI KKEQEEV AAK EDALKA RKKL LNNTH DFLED LIFR KQKIKE LMD HRAKVLS DL ENKYKKEK E ALEKETRGK ILTAKSKAYG DLEQALKDN P LYRKLLPN PY AYVLNQE TFT KEDRER LSYY YPQVK ...String: MLAKIVFSSL VAFGVLSAN V EQFGSFFN EI KKEQEEV AAK EDALKA RKKL LNNTH DFLED LIFR KQKIKE LMD HRAKVLS DL ENKYKKEK E ALEKETRGK ILTAKSKAYG DLEQALKDN P LYRKLLPN PY AYVLNQE TFT KEDRER LSYY YPQVK TSSIF KKTT ATTKDK AQA LLQMGVF SL DEEQNKKA S RLALSYKQA IEEYSNNVSN LLSRKELDN I DYYLQLER NK FDSKAKD IAQ KATNTL IFNS ERLAF SMAID KINE KYLRGY EAF SNLLKNV KD DVELNTLT K NFTNQKLSF AQKQKLCLLV LDSFNFDTQ S KKSILKKT NE YNIFVDS DPM MSDKTT MQKE HYKIF NFFKT VVSA YRNNVA KNN PFE |
-Macromolecule #3: Type IV secretion system apparatus protein CagT
| Macromolecule | Name: Type IV secretion system apparatus protein CagT / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Helicobacter pylori 26695 (bacteria) / Strain: 26695 Helicobacter pylori 26695 (bacteria) / Strain: 26695 |
| Sequence | String: MLAKIVFSSL VAFGVLSAN V EQFGSFFN EI KKEQEEV AAK EDALKA RKKL LNNTH DFLED LIFR KQKIKE LMD HRAKVLS DL ENKYKKEK E ALEKETRGK ILTAKSKAYG DLEQALKDN P LYRKLLPN PY AYVLNQE TFT KEDRER LSYY YPQVK ...String: MLAKIVFSSL VAFGVLSAN V EQFGSFFN EI KKEQEEV AAK EDALKA RKKL LNNTH DFLED LIFR KQKIKE LMD HRAKVLS DL ENKYKKEK E ALEKETRGK ILTAKSKAYG DLEQALKDN P LYRKLLPN PY AYVLNQE TFT KEDRER LSYY YPQVK TSSIF KKTT ATTKDK AQA LLQMGVF SL DEEQNKKA S RLALSYKQA IEEYSNNVSN LLSRKELDN I DYYLQLER NK FDSKAKD IAQ KATNTL IFNS ERLAF SMAID KINE KYLRGY EAF SNLLKNV KD DVELNTLT K NFTNQKLSF AQKQKLCLLV LDSFNFDTQ S KKSILKKT NE YNIFVDS DPM MSDKTT MQKE HYKIF NFFKT VVSA YRNNVA KNN PFE |
-Macromolecule #4: Type IV secretion system apparatus protein CagX
| Macromolecule | Name: Type IV secretion system apparatus protein CagX / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Helicobacter pylori 26695 (bacteria) / Strain: 26695 Helicobacter pylori 26695 (bacteria) / Strain: 26695 |
| Sequence | String: MKLRASVLIG ATILCLILSA CSNYAKKVVK QKNHVYTPVY NELIEKYSEI PLNDKLKDTP FMVQVKLPN YKDYLLDNKQ VVLTFKLVHH SKKITLIGDA NKILQYKNYF QANGARSDID F YLQPTLNQ KGVVMIASNY NDNPNSKEKP QTFDVLQGSQ PMLGANTKNL ...String: MKLRASVLIG ATILCLILSA CSNYAKKVVK QKNHVYTPVY NELIEKYSEI PLNDKLKDTP FMVQVKLPN YKDYLLDNKQ VVLTFKLVHH SKKITLIGDA NKILQYKNYF QANGARSDID F YLQPTLNQ KGVVMIASNY NDNPNSKEKP QTFDVLQGSQ PMLGANTKNL HGYDVSGANN KQ VINEVAR EKAQLEKINQ YYKTLLQDKE QEYTTRKNNQ REILETLSNR AGYQMRQNVI SSE IFKNGN LNMQAKEEEV REKLQEEREN EYLRNQIRSL LSGK |
-Macromolecule #5: Cag pathogenicity island protein (Cag7)
| Macromolecule | Name: Cag pathogenicity island protein (Cag7) / type: protein_or_peptide / ID: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Helicobacter pylori 26695 (bacteria) / Strain: 26695 Helicobacter pylori 26695 (bacteria) / Strain: 26695 |
| Sequence | String: MGQAFFKKIV GCFCLGYLFL SSAIEAAALD IKNFNRGRVK VVNKKIAYLG DEKPITIWTS LDNVTVIQL EKDETISYIT TGFNKGWSIV PNSNHIFIQP KSVKSNLMFE KEAVNFALMT R DYQEFLKT KKLIVDAPDP KELEEQKKAL EKEKEAKEQA QKAQKDKREK ...String: MGQAFFKKIV GCFCLGYLFL SSAIEAAALD IKNFNRGRVK VVNKKIAYLG DEKPITIWTS LDNVTVIQL EKDETISYIT TGFNKGWSIV PNSNHIFIQP KSVKSNLMFE KEAVNFALMT R DYQEFLKT KKLIVDAPDP KELEEQKKAL EKEKEAKEQA QKAQKDKREK RKEERAKNRA NL ENLTNAM SNPQNLSNNK NLSEFIKQQR ENELDQMERL EDMQEQAQAN ALKQIEELNK KQA EETIKQ RAKDKINIKT DKPQKSPEDN SIELSPSDSA WRTNLVVRTN KALYQFILRI AQKD NFASA YLTVKLEYPQ RHEVSSVIEE ELKKREEAKR QKELIKQENL NTTAYINRVM MASNE QIIN KEKIREEKQK IILDQAKALE TQYVHNALKR NPVPRNYNYY QAPEKRSKHI MPSEIF DDG TFTYFGFKNI TLQPAIFVVQ PDGKLSMTDA AIDPNMTNSG LRWYRVNEIA EKFKLIK DK ALVTVINKGY GKNPLTKNYN IKNYGELERV IKKLPLVRDK |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL |
|---|---|
| Buffer | pH: 7 |
| Vitrification | Cryogen name: NITROGEN |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)