[English] 日本語
 Yorodumi
Yorodumi- EMDB-41082: Cryo-EM structure of human Anion Exchanger 1 bound to 4,4'-Diisot... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human Anion Exchanger 1 bound to 4,4'-Diisothiocyanatostilbene-2,2'-Disulfonic Acid (DIDS) | ||||||||||||
 Map data Map data | structure of human Anion Exchanger 1 bound to 4,4'-Diisothiocyanatostilbene-2,2'-Disulfonic Acid (DIDS) | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords |  Transmembrane / Transmembrane /  TRANSPORT PROTEIN TRANSPORT PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationpH elevation / Defective SLC4A1 causes hereditary spherocytosis type 4 (HSP4), distal renal tubular acidosis (dRTA) and dRTA with hemolytic anemia (dRTA-HA) / negative regulation of urine volume / Bicarbonate transporters / intracellular monoatomic ion homeostasis /  ankyrin-1 complex / plasma membrane phospholipid scrambling / monoatomic anion transmembrane transporter activity / chloride:bicarbonate antiporter activity / solute:inorganic anion antiporter activity ...pH elevation / Defective SLC4A1 causes hereditary spherocytosis type 4 (HSP4), distal renal tubular acidosis (dRTA) and dRTA with hemolytic anemia (dRTA-HA) / negative regulation of urine volume / Bicarbonate transporters / intracellular monoatomic ion homeostasis / ankyrin-1 complex / plasma membrane phospholipid scrambling / monoatomic anion transmembrane transporter activity / chloride:bicarbonate antiporter activity / solute:inorganic anion antiporter activity ...pH elevation / Defective SLC4A1 causes hereditary spherocytosis type 4 (HSP4), distal renal tubular acidosis (dRTA) and dRTA with hemolytic anemia (dRTA-HA) / negative regulation of urine volume / Bicarbonate transporters / intracellular monoatomic ion homeostasis /  ankyrin-1 complex / plasma membrane phospholipid scrambling / monoatomic anion transmembrane transporter activity / chloride:bicarbonate antiporter activity / solute:inorganic anion antiporter activity / bicarbonate transmembrane transporter activity / bicarbonate transport / monoatomic anion transport / chloride transmembrane transporter activity / chloride transport / erythrocyte development / negative regulation of glycolytic process through fructose-6-phosphate / ankyrin-1 complex / plasma membrane phospholipid scrambling / monoatomic anion transmembrane transporter activity / chloride:bicarbonate antiporter activity / solute:inorganic anion antiporter activity / bicarbonate transmembrane transporter activity / bicarbonate transport / monoatomic anion transport / chloride transmembrane transporter activity / chloride transport / erythrocyte development / negative regulation of glycolytic process through fructose-6-phosphate /  ankyrin binding / ankyrin binding /  hemoglobin binding / cortical cytoskeleton / protein-membrane adaptor activity / chloride transmembrane transport / protein localization to plasma membrane / hemoglobin binding / cortical cytoskeleton / protein-membrane adaptor activity / chloride transmembrane transport / protein localization to plasma membrane /  regulation of intracellular pH / Erythrocytes take up oxygen and release carbon dioxide / Erythrocytes take up carbon dioxide and release oxygen / transmembrane transport / cytoplasmic side of plasma membrane / Z disc / regulation of intracellular pH / Erythrocytes take up oxygen and release carbon dioxide / Erythrocytes take up carbon dioxide and release oxygen / transmembrane transport / cytoplasmic side of plasma membrane / Z disc /  blood coagulation / basolateral plasma membrane / blood microparticle / protein homodimerization activity / extracellular exosome / blood coagulation / basolateral plasma membrane / blood microparticle / protein homodimerization activity / extracellular exosome /  membrane / membrane /  plasma membrane plasma membraneSimilarity search - Function | ||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.95 Å cryo EM / Resolution: 2.95 Å | ||||||||||||
 Authors Authors | Capper MJ / Zilberg G / Mathiharan YK / Yang S / Stone AC / Wacker D | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2023 Journal: Nat Struct Mol Biol / Year: 2023Title: Substrate binding and inhibition of the anion exchanger 1 transporter. Authors: Michael J Capper / Shifan Yang / Alexander C Stone / Sezen Vatansever / Gregory Zilberg / Yamuna Kalyani Mathiharan / Raul Habib / Keino Hutchinson / Yihan Zhao / Avner Schlessinger / Mihaly ...Authors: Michael J Capper / Shifan Yang / Alexander C Stone / Sezen Vatansever / Gregory Zilberg / Yamuna Kalyani Mathiharan / Raul Habib / Keino Hutchinson / Yihan Zhao / Avner Schlessinger / Mihaly Mezei / Roman Osman / Bin Zhang / Daniel Wacker /  Abstract: Anion exchanger 1 (AE1), a member of the solute carrier (SLC) family, is the primary bicarbonate transporter in erythrocytes, regulating pH levels and CO transport between lungs and tissues. Previous ...Anion exchanger 1 (AE1), a member of the solute carrier (SLC) family, is the primary bicarbonate transporter in erythrocytes, regulating pH levels and CO transport between lungs and tissues. Previous studies characterized its role in erythrocyte structure and provided insight into transport regulation. However, key questions remain regarding substrate binding and transport, mechanisms of drug inhibition and modulation by membrane components. Here we present seven cryo-EM structures in apo, bicarbonate-bound and inhibitor-bound states. These, combined with uptake and computational studies, reveal important molecular features of substrate recognition and transport, and illuminate sterol binding sites, to elucidate distinct inhibitory mechanisms of research chemicals and prescription drugs. We further probe the substrate binding site via structure-based ligand screening, identifying an AE1 inhibitor. Together, our findings provide insight into mechanisms of solute carrier transport and inhibition. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41082.map.gz emd_41082.map.gz | 30.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41082-v30.xml emd-41082-v30.xml emd-41082.xml emd-41082.xml | 22.2 KB 22.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_41082.png emd_41082.png | 117.1 KB | ||
| Filedesc metadata |  emd-41082.cif.gz emd-41082.cif.gz | 7.1 KB | ||
| Others |  emd_41082_additional_1.map.gz emd_41082_additional_1.map.gz emd_41082_half_map_1.map.gz emd_41082_half_map_1.map.gz emd_41082_half_map_2.map.gz emd_41082_half_map_2.map.gz | 59.8 MB 59 MB 59 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41082 http://ftp.pdbj.org/pub/emdb/structures/EMD-41082 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41082 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41082 | HTTPS FTP |
-Related structure data
| Related structure data |  8t6vMC  7ty4C  7ty6C  7ty7C  7ty8C  7tyaC  8t6uC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_41082.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41082.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | structure of human Anion Exchanger 1 bound to 4,4'-Diisothiocyanatostilbene-2,2'-Disulfonic Acid (DIDS) | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.083 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Auto-sharpened
| File | emd_41082_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Auto-sharpened | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map 1
| File | emd_41082_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map 2
| File | emd_41082_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Dimeric anion exchanger 1 (SLC4A1)
| Entire | Name: Dimeric anion exchanger 1 (SLC4A1) |
|---|---|
| Components |
|
-Supramolecule #1: Dimeric anion exchanger 1 (SLC4A1)
| Supramolecule | Name: Dimeric anion exchanger 1 (SLC4A1) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 203 KDa |
-Macromolecule #1: Band 3 anion transport protein
| Macromolecule | Name: Band 3 anion transport protein / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 58.254105 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: DDPLQQTGQL FGGLVRDIRR RYPYYLSDIT DAFSPQVLAA VIFIYFAALS PAITFGGLLG EKTRNQMGVS ELLISTAVQG ILFALLGAQ PLLVVGFSGP LLVFEEAFFS FCETNGLEYI VGRVWIGFWL ILLVVLVVAF EGSFLVRFIS RYTQEIFSFL I SLIFIYET ...String: DDPLQQTGQL FGGLVRDIRR RYPYYLSDIT DAFSPQVLAA VIFIYFAALS PAITFGGLLG EKTRNQMGVS ELLISTAVQG ILFALLGAQ PLLVVGFSGP LLVFEEAFFS FCETNGLEYI VGRVWIGFWL ILLVVLVVAF EGSFLVRFIS RYTQEIFSFL I SLIFIYET FSKLIKIFQD HPLQKTYNYN VLMVPKPQGP LPNTALLSLV LMAGTFFFAM MLRKFKNSSY FPGKLRRVIG DF GVPISIL IMVLVDFFIQ DTYTQKLSVP DGFKVSNSSA RGWVIHPLGL RSEFPIWMMF ASALPALLVF ILIFLESQIT TLI VSKPER KMVKGSGFHL DLLLVVGMGG VAALFGMPWL SATTVRSVTH ANALTVMGKA STPGAAAQIQ EVKEQRISGL LVAV LVGLS ILMEPILSRI PLAVLFGIFL YMGVTSLSGI QLFDRILLLF KPPKYHPDVP YVKRVKTWRM HLFTGIQIIC LAVLW VVKS TPASLALPFV LILTVPLRRV LLPLIFRNVE LQCLDADDA UniProtKB:  Band 3 anion transport protein Band 3 anion transport protein |
-Macromolecule #3: 4,4'-Diisothiocyano-2,2'-stilbenedisulfonic acid
| Macromolecule | Name: 4,4'-Diisothiocyano-2,2'-stilbenedisulfonic acid / type: ligand / ID: 3 / Number of copies: 2 / Formula: 4DS |
|---|---|
| Molecular weight | Theoretical: 454.52 Da |
| Chemical component information |  ChemComp-4DS: |
-Macromolecule #4: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 4 / Number of copies: 6 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Macromolecule #5: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 5 / Number of copies: 2 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information | 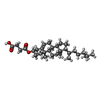 ChemComp-Y01: |
-Macromolecule #6: 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE
| Macromolecule | Name: 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE / type: ligand / ID: 6 / Number of copies: 2 / Formula: PC1 |
|---|---|
| Molecular weight | Theoretical: 790.145 Da |
| Chemical component information |  ChemComp-PC1: |
-Macromolecule #7: water
| Macromolecule | Name: water / type: ligand / ID: 7 / Number of copies: 64 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 285 K / Instrument: LEICA EM GP Details: Blot force 3 for 3-5 seconds was used and subsequent grids were screened for ice thickness prior to data collection. | ||||||||||||
| Details | The sample was monodisperse following gel filtration and immediately concentrated for grid preparation |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 81000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 5910 / Average electron dose: 52.09 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Particle selection | Number selected: 5921308 |
|---|---|
| Startup model | Type of model: NONE |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC |
| Final 3D classification | Number classes: 3 / Software - Name: cryoSPARC Details: Multiple rounds of 3D classification to remove particles |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC |
| Final reconstruction | Number classes used: 1 / Applied symmetry - Point group: C2 (2 fold cyclic ) / Resolution.type: BY AUTHOR / Resolution: 2.95 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC / Number images used: 914784 ) / Resolution.type: BY AUTHOR / Resolution: 2.95 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC / Number images used: 914784 |
 Movie
Movie Controller
Controller









 Z
Z Y
Y X
X