+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of DSR2 (H171A)-tube-NAD+ (partial) complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Phage defense proteins /  ANTIVIRAL PROTEIN ANTIVIRAL PROTEIN | |||||||||
| Function / homology | SIR2-like domain / SIR2-like domain / DHS-like NAD/FAD-binding domain superfamily / Uncharacterized protein / SIR2-like domain-containing protein Function and homology information Function and homology information | |||||||||
| Biological species |   Bacillus subtilis (bacteria) / Bacillus subtilis (bacteria) /  Bacillus phage SPR (virus) Bacillus phage SPR (virus) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.0 Å cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Zhang JT / Jia N / Liu XY | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structural basis for phage-mediated activation and repression of bacterial DSR2 anti-phage defense system. Authors: Jun-Tao Zhang / Xiao-Yu Liu / Zhuolin Li / Xin-Yang Wei / Xin-Yi Song / Ning Cui / Jirui Zhong / Hongchun Li / Ning Jia /  Abstract: Silent information regulator 2 (Sir2) proteins typically catalyze NAD-dependent protein deacetylation. The recently identified bacterial Sir2 domain-containing protein, defense-associated sirtuin 2 ...Silent information regulator 2 (Sir2) proteins typically catalyze NAD-dependent protein deacetylation. The recently identified bacterial Sir2 domain-containing protein, defense-associated sirtuin 2 (DSR2), recognizes the phage tail tube and depletes NAD to abort phage propagation, which is counteracted by the phage-encoded DSR anti-defense 1 (DSAD1), but their molecular mechanisms remain unclear. Here, we determine cryo-EM structures of inactive DSR2 in its apo form, DSR2-DSAD1 and DSR2-DSAD1-NAD, as well as active DSR2-tube and DSR2-tube-NAD complexes. DSR2 forms a tetramer with its C-terminal sensor domains (CTDs) in two distinct conformations: CTD or CTD. Monomeric, rather than oligomeric, tail tube proteins preferentially bind to CTD and activate Sir2 for NAD hydrolysis. DSAD1 binding to CTD allosterically inhibits tube binding and tube-mediated DSR2 activation. Our findings provide mechanistic insight into DSR2 assembly, tube-mediated DSR2 activation, and DSAD1-mediated inhibition and NAD substrate catalysis in bacterial DSR2 anti-phage defense systems. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37923.map.gz emd_37923.map.gz | 567.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37923-v30.xml emd-37923-v30.xml emd-37923.xml emd-37923.xml | 15.7 KB 15.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_37923.png emd_37923.png | 85.9 KB | ||
| Masks |  emd_37923_msk_1.map emd_37923_msk_1.map | 600.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-37923.cif.gz emd-37923.cif.gz | 6.1 KB | ||
| Others |  emd_37923_half_map_1.map.gz emd_37923_half_map_1.map.gz emd_37923_half_map_2.map.gz emd_37923_half_map_2.map.gz | 558.1 MB 558.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37923 http://ftp.pdbj.org/pub/emdb/structures/EMD-37923 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37923 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37923 | HTTPS FTP |
-Related structure data
| Related structure data |  8wycMC  8wy8C  8wy9C  8wyaC  8wybC  8wydC  8wyeC  8wyfC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37923.map.gz / Format: CCP4 / Size: 600.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37923.map.gz / Format: CCP4 / Size: 600.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.827 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_37923_msk_1.map emd_37923_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
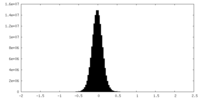
| Density Histograms |
-Half map: #2
| File | emd_37923_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
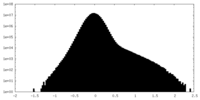
| Density Histograms |
-Half map: #1
| File | emd_37923_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : CryoEM structure of DSR2 (H171A)-Tube NAD+ (half) complex
| Entire | Name: CryoEM structure of DSR2 (H171A)-Tube NAD+ (half) complex |
|---|---|
| Components |
|
-Supramolecule #1: CryoEM structure of DSR2 (H171A)-Tube NAD+ (half) complex
| Supramolecule | Name: CryoEM structure of DSR2 (H171A)-Tube NAD+ (half) complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Bacillus subtilis (bacteria) Bacillus subtilis (bacteria) |
-Macromolecule #1: SIR2-like domain-containing protein
| Macromolecule | Name: SIR2-like domain-containing protein / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Bacillus subtilis (bacteria) Bacillus subtilis (bacteria) |
| Molecular weight | Theoretical: 118.568727 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MVKVDLESKR YGEKLKEVFL MLDNNVVECI KEITESSRNG KLVFFVGAGV STLSDYPQWW RLVDKYHEEL YGSPKKGNYS SDEYLRIPQ IFYNVKGEMA FDGILKDFFQ VDKPTNPIHD KILAMNPAHV ITTNYDNLID TACWKRGKYF SVISAEEDVA N ATSSRYLL ...String: MVKVDLESKR YGEKLKEVFL MLDNNVVECI KEITESSRNG KLVFFVGAGV STLSDYPQWW RLVDKYHEEL YGSPKKGNYS SDEYLRIPQ IFYNVKGEMA FDGILKDFFQ VDKPTNPIHD KILAMNPAHV ITTNYDNLID TACWKRGKYF SVISAEEDVA N ATSSRYLL KVAGDFRKGF KGENVVLKED DYLNYDQNYP LISNLMKTII ATHTIVFIGY GLGDYNINML LNWVRKLQKD SF HKPFFIR TDPSPIENET LIYYENKGLR IIDAASLIDS NEYDYLERYS AVMDLLIESQ ENKFITKDDE VIDYIYGKIS PLF ALQYIR KIDLKHVFEY DYHFEVNGTV VRHKNKGFGY MERFFELKES CDERSKLSKK QYERFNALFN FFEKNGVICM AKDA GTLNT SIEINSLAYH GKYDVMKKFI EEQSVSIEDD YKKAFFLACL GRWEESYDLY SNIILNSIDE SNGCVYYLSQ INRYR IYQS ITQAVTQFNG LGLLTFGRHY KPFTDEFLAR IEREMTNFNI DDLFNGMPFE FQKKYKILEF LSDNQFLYDD TVKLFE LTN KVRSEMSEGS YSFGMSSDIV VLLRLYDNLR FLYENCLWSV SFHEFHQYIR NSMSLLIEKA EYERTRDIDE LGFSFFG KK SGFFMEYYDF VNISRHFKID DIKNLERSCS IDKIRFGEQE KIEEYLVGIA EEITKQFSAN GMNVVFYTQF ISEAKAAL Y FAKYVKLSEE GLGKIVKALL FYFPERDLDI GKRYVWLERL TKCNELPKSI ISIIDDFLVL QAEKHIDQNY SEVSSNGLY SRDYGALIKH FEKNFISKRL SEITLCLTQD KQKQIDFLFK LLPLLSTNAK SHLLSFKSVE NINDLMNGIR IGLIDEFTPE HEELIIEYL ETRKVNYIVE KEKGIQTFSS NDYMSTFGIW YFLEEINNSK MEEFIGMDDQ YDFFVDPENF DYKKFIPSWL K NYNDKLLG KIAGNKHMKH HVIEVLKERV KNSNDKRYLE ILMNYFI UniProtKB: SIR2-like domain-containing protein |
-Macromolecule #2: Bacillus phage SPR Tube protein
| Macromolecule | Name: Bacillus phage SPR Tube protein / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Bacillus phage SPR (virus) Bacillus phage SPR (virus) |
| Molecular weight | Theoretical: 29.304701 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MKTVIQDTAD VYFKRKSDGK LVFTAEAQTA SFSQAISEEK LRGGIGNKPL YILKSEKEIN LTVKNAFFDL EWLAMTQGET IQEETKVKV FDREHGLIVD DTNKVTLKGK PVSDVTFYNK KGLTYKIAVS TDGTYTIPTA FAAAKDKLTA VYQIEKVGRR L AIKASKFS ...String: MKTVIQDTAD VYFKRKSDGK LVFTAEAQTA SFSQAISEEK LRGGIGNKPL YILKSEKEIN LTVKNAFFDL EWLAMTQGET IQEETKVKV FDREHGLIVD DTNKVTLKGK PVSDVTFYNK KGLTYKIAVS TDGTYTIPTA FAAAKDKLTA VYQIEKVGRR L AIKASKFS ERYEVEYRTI AYNPDTEEVY SDIYIQFPNV SPSGEFEMSL ENGNALAPEI KFEALADTDT DEMAVVIEAS RD ENTAAPV EDTTGSTQSS DLGGTTE UniProtKB: Uncharacterized protein |
-Macromolecule #3: NICOTINAMIDE-ADENINE-DINUCLEOTIDE
| Macromolecule | Name: NICOTINAMIDE-ADENINE-DINUCLEOTIDE / type: ligand / ID: 3 / Number of copies: 4 / Formula: NAD |
|---|---|
| Molecular weight | Theoretical: 663.425 Da |
| Chemical component information |  ChemComp-NAD: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: RANDOM ASSIGNMENT |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.0 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 85495 |
 Movie
Movie Controller
Controller













 Z
Z Y
Y X
X