[English] 日本語
 Yorodumi
Yorodumi- EMDB-37389: cryo-EM structure of native mastigonemes isolated from Chlamydomo... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryo-EM structure of native mastigonemes isolated from Chlamydomonas reinhardtii at 3.0 angstrom resolution | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Cilia / arabinoglycan / Cilia / arabinoglycan /  PKD2 / PPII helix / PKD2 / PPII helix /  mechanosensation. / mechanosensation. /  MEMBRANE PROTEIN MEMBRANE PROTEIN | |||||||||
| Function / homology | Tyrosine-protein kinase ephrin type A/B receptor-like / Tyrosine-protein kinase ephrin type A/B receptor-like / Putative ephrin-receptor like / Growth factor receptor cysteine-rich domain superfamily / Tyrosine-protein kinase ephrin type A/B receptor-like domain-containing protein Function and homology information Function and homology information | |||||||||
| Biological species |   Chlamydomonas reinhardtii (plant) / synthetic construct (others) Chlamydomonas reinhardtii (plant) / synthetic construct (others) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.0 Å cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Huang J / Tao H / Chen J / Pan J / Yan C / Yan N | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2024 Journal: Cell / Year: 2024Title: Structure-guided discovery of protein and glycan components in native mastigonemes. Authors: Junhao Huang / Hui Tao / Jikun Chen / Yang Shen / Jianlin Lei / Junmin Pan / Chuangye Yan / Nieng Yan /  Abstract: Mastigonemes, the hair-like lateral appendages lining cilia or flagella, participate in mechanosensation and cellular motion, but their constituents and structure have remained unclear. Here, we ...Mastigonemes, the hair-like lateral appendages lining cilia or flagella, participate in mechanosensation and cellular motion, but their constituents and structure have remained unclear. Here, we report the cryo-EM structure of native mastigonemes isolated from Chlamydomonas at 3.0 Å resolution. The long stem assembles as a super spiral, with each helical turn comprising four pairs of anti-parallel mastigoneme-like protein 1 (Mst1). A large array of arabinoglycans, which represents a common class of glycosylation in plants and algae, is resolved surrounding the type II poly-hydroxyproline (Hyp) helix in Mst1. The EM map unveils a mastigoneme axial protein (Mstax) that is rich in heavily glycosylated Hyp and contains a PKD2-like transmembrane domain (TMD). Mstax, with nearly 8,000 residues spanning from the intracellular region to the distal end of the mastigoneme, provides the framework for Mst1 assembly. Our study provides insights into the complexity of protein and glycan interactions in native bio-architectures. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37389.map.gz emd_37389.map.gz | 483.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37389-v30.xml emd-37389-v30.xml emd-37389.xml emd-37389.xml | 23.6 KB 23.6 KB | Display Display |  EMDB header EMDB header |
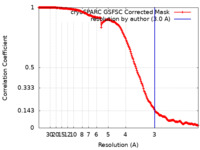
| FSC (resolution estimation) |  emd_37389_fsc.xml emd_37389_fsc.xml | 16.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_37389.png emd_37389.png | 80.7 KB | ||
| Masks |  emd_37389_msk_1.map emd_37389_msk_1.map | 512 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-37389.cif.gz emd-37389.cif.gz | 7.3 KB | ||
| Others |  emd_37389_additional_1.map.gz emd_37389_additional_1.map.gz emd_37389_additional_2.map.gz emd_37389_additional_2.map.gz emd_37389_additional_3.map.gz emd_37389_additional_3.map.gz emd_37389_half_map_1.map.gz emd_37389_half_map_1.map.gz emd_37389_half_map_2.map.gz emd_37389_half_map_2.map.gz | 483.2 MB 483.2 MB 483.4 MB 475.2 MB 475.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37389 http://ftp.pdbj.org/pub/emdb/structures/EMD-37389 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37389 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37389 | HTTPS FTP |
-Related structure data
| Related structure data |  8wa2MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37389.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37389.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.0825 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_37389_msk_1.map emd_37389_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_37389_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #2
| File | emd_37389_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #3
| File | emd_37389_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_37389_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_37389_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : mastigoneme
| Entire | Name: mastigoneme |
|---|---|
| Components |
|
-Supramolecule #1: mastigoneme
| Supramolecule | Name: mastigoneme / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Chlamydomonas reinhardtii (plant) Chlamydomonas reinhardtii (plant) |
-Macromolecule #1: Mst1
| Macromolecule | Name: Mst1 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Chlamydomonas reinhardtii (plant) Chlamydomonas reinhardtii (plant) |
| Molecular weight | Theoretical: 206.316203 KDa |
| Sequence | String: MAMPRHGRRP ARCSSNRASQ WLLLVLGVAL AASPRFLLLV EAATYTLTLD KLGPTVNPTT SDAVTFTATV ASPDSTTAVF FTLDYGDGV TAETTRTTTG ALSTTPTANL VSAGTYTVTY ASIGTKFVTL RLYDSAVAPG VLLASKTVPI YVEDSTLTAT L LQSGVPRL ...String: MAMPRHGRRP ARCSSNRASQ WLLLVLGVAL AASPRFLLLV EAATYTLTLD KLGPTVNPTT SDAVTFTATV ASPDSTTAVF FTLDYGDGV TAETTRTTTG ALSTTPTANL VSAGTYTVTY ASIGTKFVTL RLYDSAVAPG VLLASKTVPI YVEDSTLTAT L LQSGVPRL NLAFSGFKGR VSSSTANRAD MWATIQLDTA PGVFESSRIF IGIAPTASTN YDFVIPDQVY NLEGAKTTVL RI YDAPVGG TLLRTFTPAA ANAVYVVDPS KYVLTLTVGP TSVTTADQVT FTQTTTEVSY SASATSPILQ WRFNWDDPSV VET PLAYPD ALTAASNFPT TATAVSSAAA STFRYTSTGS KNARLRLYDG ANNVIAEKIV VITVSNAGYT LALAKTTADP VTTD DTIAF SAGAKHLSST SQVWWTIDYG AGESSPRTAL TMTNVGAAAP NAIASLSNQY TSGGTKLATL RIYDRDGVGA NTGLL LAST TVTFTVTPVL YALESAVEPF SPIATVAAKW SFRIQRSKAT PAGVTESIKC AFFGADTGTA PADLAAWLTA ANGAGG LTA TILPSSIPSD IISFTRTYAA AAASLQGKLQ CFIGSTPLWD PYYPTPVFQV LAAAPTYTLS ASVTPAVVPV DTATLWT YN IIRSVPVPAG GPSLPILCSF WDGKTGAAPT TDAGWAALAG SANGKGTSMA PGSTTATCSF TPSYSTTGTA TPTLQLIQ N SFALDAATTV GFLSPVYTAP AFATVTAASY TISSYLNPVT PVAGGAAAVW RIVITRNAAV TASAKTLTCQ MPDNGQGGS PADVTADIAV GGTTTVCVFS IAGYTTATPG PYFATVNVVD GAVTTSHITK NFTVLASGTT APTYAVTSVV SPATPVKVST PVTYTFTIT RTTAVPAGGI PQPIICEFFN GEGTAPASAA AYWRVSTTIP DADTVVAVMA PGETTTTCTF TTYYTTVSAG G FTAKLMVF GESATAAPLL TSLSVTPSQL LAAVHSFATP MVVAAAVVAV ESTTISPNYN PTTPYTNIPT YFTFTLLRDP PV PPSASSG VQFACALYTG QNVNPASAPS AITDAVYKTF TDVTTAVATD ANYFADQQLR VVTMAPGTGR VSCTFPTLYA AAG PFSPKF FVFEYASSTV GANALAVADT VTSLTSFTTQ AAPTFITGPT NVPQRVPLPK GFRTTCFDGY ELIFSNDNYT NGVR VAVDA YPYPVGQCRK CPGGTATMDG YRCIPCPSGY WSNEGARECT ACPAGTIAKP AALTARAKYS IDPTTYHFVT HLAMG PESC KKCPKGYFQP NIAGTVCLPC PSGFVSTSGA TGCTACSEGT YHTDGVGTTT PGEATSLDTT DTFGSIYPII PNTCRQ CPA NTYLPLRGQA AIASMNLAAV SSATPCRPCE DGTWSKAGAA GCQKCPPGTY RNTWFSGQLG SPFITADGVP VATTLTE LG SGCSQCPPGT YAPTFGMSVC LPCPAGTFAS APGATACQQC KPGTNSLMGD RTQQMALVVT NAANDFPALR AYTISGMV A GPAYAKPIVT GPDTNFFMAG KSETCSTNLP GYYTDVDGLP IQLPCKPGTF MPFDTATANL LDTGLTVDGT QCYTCQTGT FNDEFSQPVC KACWSGSFAS KRGLPTCEIA QPGTFTNVAA AANATFNTAT LIPTGLVKGA QAPTPCGMGY FQSSAETTTC TACAVGTYA DQAGLAACKP CQPGRYQNSI GQRVCKPCDM GTYSRYGGEL CTKCPAGTVA SKTGSSQCTP CAAGFYANAP D SATSCRAC PRGYYGPYSG AYADNLGDEF EGPRGCYKCP YDFFADRPGV RQCTACPPLD LGGGNLVEQC TEDLGSQRCK PC SLLSKPK TARTEQS(HYP)(HYP)(HYP) (HYP)S(HYP)S(HYP)(HYP)(HYP)(HYP)(HYP)(HYP) (HYP)S (HYP)R(HYP)(HYP)S(HYP)N (HYP)(HYP)S(HYP)R(HYP)(HYP)S(HYP)A (HYP)(HYP)S(HYP)N(HYP) (HYP)(HYP)T S(HYP)(HYP)(HYP)S(HYP)(HYP)(HYP)S(HYP) (HYP)(HYP)(HYP)R(HYP)(HYP)(HYP) (HYP)(HYP) (HYP)(HYP)(HYP)(HYP)S(HYP)(HYP)(HYP)(HYP)N RS(HYP)(HYP)(HYP)(HYP)(HYP) (HYP)A SSAINPGGGV NQNGDPVGHR RAILSLMEDE DAEAAQEEQA IVDVDAEMQP QDDE UniProtKB: Tyrosine-protein kinase ephrin type A/B receptor-like domain-containing protein |
-Macromolecule #2: Mstax
| Macromolecule | Name: Mstax / type: protein_or_peptide / ID: 2 Details: Our Mstax sequence in the structure (chain G, H and I) represents the average sequence resulting from the statistical analysis of a segment of 34 repeated sequences in Mstax. Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 6.239464 KDa |
| Sequence | String: (HYP)(HYP)(HYP)(HYP)(HYP)(HYP)(HYP)(HYP)(HYP)(HYP) (HYP)GT(HYP)DQ(HYP)AA(HYP) AA (HYP)AA(HYP)AA(HYP) AA(HYP)(HYP)(HYP)AL(HYP)GA (HYP)(HYP)(HYP)(HYP)(HYP)(HYP)(HYP) (HYP)(HYP) (HYP)(HYP)(HYP) ...String: (HYP)(HYP)(HYP)(HYP)(HYP)(HYP)(HYP)(HYP)(HYP)(HYP) (HYP)GT(HYP)DQ(HYP)AA(HYP) AA (HYP)AA(HYP)AA(HYP) AA(HYP)(HYP)(HYP)AL(HYP)GA (HYP)(HYP)(HYP)(HYP)(HYP)(HYP)(HYP) (HYP)(HYP) (HYP)(HYP)(HYP)(HYP)GV(HYP)(HYP)AA AAAAAA |
-Macromolecule #25: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 25 / Number of copies: 6 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #26: alpha-L-arabinofuranose
| Macromolecule | Name: alpha-L-arabinofuranose / type: ligand / ID: 26 / Number of copies: 20 / Formula: AHR |
|---|---|
| Molecular weight | Theoretical: 150.13 Da |
| Chemical component information | 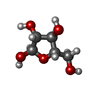 ChemComp-AHR: |
-Macromolecule #27: alpha-D-galactopyranose
| Macromolecule | Name: alpha-D-galactopyranose / type: ligand / ID: 27 / Number of copies: 62 / Formula: GLA |
|---|---|
| Molecular weight | Theoretical: 180.156 Da |
| Chemical component information | 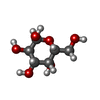 ChemComp-GLA: |
-Macromolecule #28: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 28 / Number of copies: 7 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm Bright-field microscopy / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z
Z Y
Y X
X