+ Open data
Open data
- Basic information
Basic information
| Entry | 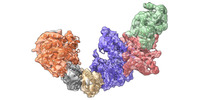 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of KEOPS complex from Arabidopsis thaliana | |||||||||
 Map data Map data | Electron density map for KEOPS complex from Arabidopsis thaliana | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | KEOPS complex / tRNA t6A synthase /  RNA-binding protein / RNA-binding protein /  TRANSFERASE TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationN(6)-L-threonylcarbamoyladenine synthase activity / EKC/KEOPS complex / tRNA threonylcarbamoyladenosine modification / tRNA processing /  non-specific serine/threonine protein kinase / non-specific serine/threonine protein kinase /  phosphorylation / protein serine/threonine kinase activity / phosphorylation / protein serine/threonine kinase activity /  ATP binding / ATP binding /  metal ion binding / metal ion binding /  nucleus / nucleus /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Arabidopsis thaliana (thale cress) Arabidopsis thaliana (thale cress) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.7 Å cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Zheng XX / Zhu L / Duan L / Zhang WH | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2024 Journal: Nucleic Acids Res / Year: 2024Title: Molecular basis of A. thaliana KEOPS complex in biosynthesizing tRNA t6A. Authors: Xinxing Zheng / Chenchen Su / Lei Duan / Mengqi Jin / Yongtao Sun / Li Zhu / Wenhua Zhang /  Abstract: In archaea and eukaryotes, the evolutionarily conserved KEOPS is composed of four core subunits-Kae1, Bud32, Cgi121 and Pcc1, and a fifth Gon7/Pcc2 that is found in fungi and metazoa. KEOPS ...In archaea and eukaryotes, the evolutionarily conserved KEOPS is composed of four core subunits-Kae1, Bud32, Cgi121 and Pcc1, and a fifth Gon7/Pcc2 that is found in fungi and metazoa. KEOPS cooperates with Sua5/YRDC to catalyze the biosynthesis of tRNA N6-threonylcarbamoyladenosine (t6A), an essential modification needed for fitness of cellular organisms. Biochemical and structural characterizations of KEOPSs from archaea, yeast and humans have determined a t6A-catalytic role for Kae1 and auxiliary roles for other subunits. However, the precise molecular workings of KEOPSs still remain poorly understood. Here, we investigated the biochemical functions of A. thaliana KEOPS and determined a cryo-EM structure of A. thaliana KEOPS dimer. We show that A. thaliana KEOPS is composed of KAE1, BUD32, CGI121 and PCC1, which adopts a conserved overall arrangement. PCC1 dimerization leads to a KEOPS dimer that is needed for an active t6A-catalytic KEOPS-tRNA assembly. BUD32 participates in direct binding of tRNA to KEOPS and modulates the t6A-catalytic activity of KEOPS via its C-terminal tail and ATP to ADP hydrolysis. CGI121 promotes the binding of tRNA to KEOPS and potentiates the t6A-catalytic activity of KEOPS. These data and findings provide insights into mechanistic understanding of KEOPS machineries. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36808.map.gz emd_36808.map.gz | 254.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36808-v30.xml emd-36808-v30.xml emd-36808.xml emd-36808.xml | 18.7 KB 18.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36808_fsc.xml emd_36808_fsc.xml | 16.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_36808.png emd_36808.png | 62 KB | ||
| Filedesc metadata |  emd-36808.cif.gz emd-36808.cif.gz | 6.3 KB | ||
| Others |  emd_36808_half_map_1.map.gz emd_36808_half_map_1.map.gz emd_36808_half_map_2.map.gz emd_36808_half_map_2.map.gz | 475.3 MB 475.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36808 http://ftp.pdbj.org/pub/emdb/structures/EMD-36808 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36808 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36808 | HTTPS FTP |
-Related structure data
| Related structure data |  8k20MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_36808.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36808.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Electron density map for KEOPS complex from Arabidopsis thaliana | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map A for KEOPS complex from Arabidopsis thaliana
| File | emd_36808_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A for KEOPS complex from Arabidopsis thaliana | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B for KEOPS complex from Arabidopsis thaliana
| File | emd_36808_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B for KEOPS complex from Arabidopsis thaliana | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : KEOPS complex from Arabidopsis thaliana
| Entire | Name: KEOPS complex from Arabidopsis thaliana |
|---|---|
| Components |
|
-Supramolecule #1: KEOPS complex from Arabidopsis thaliana
| Supramolecule | Name: KEOPS complex from Arabidopsis thaliana / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 Details: The sample contains KAE1, BUD32, CGI121 and PCC1, which form a KEOPS complex. |
|---|---|
| Source (natural) | Organism:   Arabidopsis thaliana (thale cress) Arabidopsis thaliana (thale cress) |
-Macromolecule #1: Probable tRNA N6-adenosine threonylcarbamoyltransferase
| Macromolecule | Name: Probable tRNA N6-adenosine threonylcarbamoyltransferase type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: N6-L-threonylcarbamoyladenine synthase |
|---|---|
| Source (natural) | Organism:   Arabidopsis thaliana (thale cress) Arabidopsis thaliana (thale cress) |
| Molecular weight | Theoretical: 38.81357 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MKKKMIAIGF EGSANKIGVG IVTLDGTILA NPRHTYITPP GHGFLPRETA HHHLDHVLPL VKSALETSQV TPEEIDCICY TKGPGMGAP LQVSAIVVRV LSQLWKKPIV AVNHCVAHIE MGRVVTGADD PVVLYVSGGN TQVIAYSEGR YRIFGETIDI A VGNCLDRF ...String: MKKKMIAIGF EGSANKIGVG IVTLDGTILA NPRHTYITPP GHGFLPRETA HHHLDHVLPL VKSALETSQV TPEEIDCICY TKGPGMGAP LQVSAIVVRV LSQLWKKPIV AVNHCVAHIE MGRVVTGADD PVVLYVSGGN TQVIAYSEGR YRIFGETIDI A VGNCLDRF ARVLKLSNDP SPGYNIEQLA KKGENFIDLP YAVKGMDVSF SGILSYIETT AEEKLKNNEC TPADLCYSLQ ET VFAMLVE ITERAMAHCD KKDVLIVGGV GCNERLQEMM RTMCSERDGK LFATDDRYCI DNGAMIAYTG LLAFVNGIET PIE DSTFTQ RFRTDEVHAV WREKEAVLLG DKKVAAN UniProtKB:  Glycoprotease 2 Glycoprotease 2 |
-Macromolecule #2: non-specific serine/threonine protein kinase
| Macromolecule | Name: non-specific serine/threonine protein kinase / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number:  non-specific serine/threonine protein kinase non-specific serine/threonine protein kinase |
|---|---|
| Source (natural) | Organism:   Arabidopsis thaliana (thale cress) Arabidopsis thaliana (thale cress) |
| Molecular weight | Theoretical: 25.170117 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MDCEENVRDE SLVLIKQGAE ARVFESTFAG RRSIVKERFS KKYRHPILDA KLTLKRLNAE ARCMTKARKL GVCTPVLYAV DTLLHSLTL EYIEGVSVKD IFLEFGTNGV VEERLDDVAA QIGAAIAKLH DGGLAHGDLT TSNMLVRSGT NQLVLIDFGL S VTSTLPED ...String: MDCEENVRDE SLVLIKQGAE ARVFESTFAG RRSIVKERFS KKYRHPILDA KLTLKRLNAE ARCMTKARKL GVCTPVLYAV DTLLHSLTL EYIEGVSVKD IFLEFGTNGV VEERLDDVAA QIGAAIAKLH DGGLAHGDLT TSNMLVRSGT NQLVLIDFGL S VTSTLPED KAVDLYVLER ALLSMHSSCG NVMDRILTAY RKSSKQWSAT FNKLAQVRQR GRKRTMIG UniProtKB:  non-specific serine/threonine protein kinase non-specific serine/threonine protein kinase |
-Macromolecule #3: At4g34412
| Macromolecule | Name: At4g34412 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Arabidopsis thaliana (thale cress) Arabidopsis thaliana (thale cress) |
| Molecular weight | Theoretical: 19.678484 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MKVFNLDRGN TLSVSLFSGV TNSKELLNSM LDGSLKLEVS FLNASLIPDI FPLLAAAQKA LISKSRDSLS TRTLHSELVY NYSGSKHIT ESLKRCGISE NTTYILAARF NASPVEMEEV AKLINGKEID LEELKTHANQ ANILKHYKIT SQELGISSLG D AIVCRIAA RDALHHHHHH UniProtKB: At4g34412 |
-Macromolecule #4: At5g53043
| Macromolecule | Name: At5g53043 / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Arabidopsis thaliana (thale cress) Arabidopsis thaliana (thale cress) |
| Molecular weight | Theoretical: 11.592994 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MAATVPATTR SDQTWDFSCN LDVSFESEEH ALIAYTSLAV DKELQPDKVR RVMSVSNNKL SVHFEAIEAR LLRASFSAFV DVLTLATRT IQEFGQKHHH HHH UniProtKB: At5g53043 |
-Macromolecule #5: FE (III) ION
| Macromolecule | Name: FE (III) ION / type: ligand / ID: 5 / Number of copies: 1 / Formula: FE |
|---|---|
| Molecular weight | Theoretical: 55.845 Da |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Details: 300 mM NaCl, 20 mM Tris-HCl pH 7.5,5 mM 2-mercaptoethanol |
|---|---|
| Vitrification | Cryogen name: ETHANE |
| Details | The sample was freshly purified by size exclusion chromatography. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 0.27 mm / Nominal defocus max: 2.7 µm / Nominal defocus min: 0.9 µm / Nominal magnification: 105000 Bright-field microscopy / Cs: 0.27 mm / Nominal defocus max: 2.7 µm / Nominal defocus min: 0.9 µm / Nominal magnification: 105000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Temperature | Min: 77.1 K / Max: 89.8 K |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Average exposure time: 3.6 sec. / Average electron dose: 64.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z
Z Y
Y X
X


















