[English] 日本語
 Yorodumi
Yorodumi- EMDB-35246: Outer shell and inner layer structures of Autographa californica ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Outer shell and inner layer structures of Autographa californica multiple nucleopolyhedrovirus (AcMNPV) | |||||||||
 Map data Map data | Outer shell and inner layer of AcMNPV | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  virus / virus /  capsid protein / capsid protein /  VIRAL PROTEIN VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |   Autographa californica multiple nucleopolyhedrovirus Autographa californica multiple nucleopolyhedrovirus | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 4.31 Å cryo EM / Resolution: 4.31 Å | |||||||||
 Authors Authors | Jia X / Gao Y / Zhang Q | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Architecture of the baculovirus nucleocapsid revealed by cryo-EM. Authors: Xudong Jia / Yuanzhu Gao / Yuxuan Huang / Linjun Sun / Siduo Li / Hongmei Li / Xueqing Zhang / Yinyin Li / Jian He / Wenbi Wu / Harikanth Venkannagari / Kai Yang / Matthew L Baker / Qinfen Zhang /   Abstract: Baculovirus Autographa californica multiple nucleopolyhedrovirus (AcMNPV) has been widely used as a bioinsecticide and a protein expression vector. Despite their importance, very little is known ...Baculovirus Autographa californica multiple nucleopolyhedrovirus (AcMNPV) has been widely used as a bioinsecticide and a protein expression vector. Despite their importance, very little is known about the structure of most baculovirus proteins. Here, we show a 3.2 Å resolution structure of helical cylindrical body of the AcMNPV nucleocapsid, composed of VP39, as well as 4.3 Å resolution structures of both the head and the base of the nucleocapsid composed of over 100 protein subunits. AcMNPV VP39 demonstrates some features of the HK97-like fold and utilizes disulfide-bonds and a set of interactions at its C-termini to mediate nucleocapsid assembly and stability. At both ends of the nucleocapsid, the VP39 cylinder is constricted by an outer shell ring composed of proteins AC104, AC142 and AC109. AC101(BV/ODV-C42) and AC144(ODV-EC27) form a C14 symmetric inner layer at both capsid head and base. In the base, these proteins interact with a 7-fold symmetric capsid plug, while a portal-like structure is seen in the central portion of head. Additionally, we propose an application of AlphaFold2 for model building in intermediate resolution density. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35246.map.gz emd_35246.map.gz | 39.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35246-v30.xml emd-35246-v30.xml emd-35246.xml emd-35246.xml | 24.4 KB 24.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35246_fsc.xml emd_35246_fsc.xml | 19.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_35246.png emd_35246.png | 117.1 KB | ||
| Masks |  emd_35246_msk_1.map emd_35246_msk_1.map | 824 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-35246.cif.gz emd-35246.cif.gz | 7.6 KB | ||
| Others |  emd_35246_additional_1.map.gz emd_35246_additional_1.map.gz emd_35246_half_map_1.map.gz emd_35246_half_map_1.map.gz emd_35246_half_map_2.map.gz emd_35246_half_map_2.map.gz | 774.3 MB 761.9 MB 761.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35246 http://ftp.pdbj.org/pub/emdb/structures/EMD-35246 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35246 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35246 | HTTPS FTP |
-Related structure data
| Related structure data |  8i8bMC  8i8aC  8i8cC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_35246.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35246.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Outer shell and inner layer of AcMNPV | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.35 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_35246_msk_1.map emd_35246_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
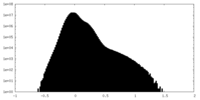
| Density Histograms |
-Additional map: additional EM map for chain I,J
| File | emd_35246_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | additional EM map for chain I,J | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 1
| File | emd_35246_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2
| File | emd_35246_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Autographa californica multiple nucleopolyhedrovirus
| Entire | Name:   Autographa californica multiple nucleopolyhedrovirus Autographa californica multiple nucleopolyhedrovirus |
|---|---|
| Components |
|
-Supramolecule #1: Autographa californica multiple nucleopolyhedrovirus
| Supramolecule | Name: Autographa californica multiple nucleopolyhedrovirus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 307456 Sci species name: Autographa californica multiple nucleopolyhedrovirus Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: Major viral capsid protein
| Macromolecule | Name: Major viral capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Autographa californica multiple nucleopolyhedrovirus Autographa californica multiple nucleopolyhedrovirus |
| Molecular weight | Theoretical: 38.991109 KDa |
| Sequence | String: MALVPVGMAP RQMRVNRCIF ASIVSFDACI TYKSPCSPDA YHDDGWFICN NHLIKRFKMS KMVLPIFDED DNQFKMTIAR HLVGNKERG IKRILIPSAT NYQDVFNLNS MMQAEQLIFH LIYNNENAVN TICDNLKYTE GFTSNTQRVI HSVYATTKSI L DTTNPNTF ...String: MALVPVGMAP RQMRVNRCIF ASIVSFDACI TYKSPCSPDA YHDDGWFICN NHLIKRFKMS KMVLPIFDED DNQFKMTIAR HLVGNKERG IKRILIPSAT NYQDVFNLNS MMQAEQLIFH LIYNNENAVN TICDNLKYTE GFTSNTQRVI HSVYATTKSI L DTTNPNTF CSRVSRDELR FFDVTNARAL RGGAGDQLFN NYSGFLQNLI RRAVAPEYLQ IDTEELRFRN CATCIIDETG LV ASVPDGP ELYNPIRSSD IMRSQPNRLQ IRNVLKFEGD TRELDRTLSG YEEYPTYVPL FLGYQIINSE NNFLRNDFIP RAN PNATLG GGAVAGPAPG VAGEAGGGIA V UniProtKB: Major viral capsid protein |
-Macromolecule #2: Viral capsid associated protein
| Macromolecule | Name: Viral capsid associated protein / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Autographa californica multiple nucleopolyhedrovirus Autographa californica multiple nucleopolyhedrovirus |
| Molecular weight | Theoretical: 79.974469 KDa |
| Sequence | String: MNDSNSLLIT RLAAQILSRN MQTVDVIVDD KTLSLEEKID TLTSMVLAVN SPPQSPPRVT SSDLAASIIK NNSKMVGNDF EMRYNVLRM AVVFVKHYPK YYNETTAGLV AEIESNLLQY QNYVNQGNYQ NIEGYDSLLN KAEECYVKID RLFKESIKKI M DDTEAFER ...String: MNDSNSLLIT RLAAQILSRN MQTVDVIVDD KTLSLEEKID TLTSMVLAVN SPPQSPPRVT SSDLAASIIK NNSKMVGNDF EMRYNVLRM AVVFVKHYPK YYNETTAGLV AEIESNLLQY QNYVNQGNYQ NIEGYDSLLN KAEECYVKID RLFKESIKKI M DDTEAFER EQEAERLRAE QTAANALLER RAQTSADDVV NRADANIPTA FSDPLPGPSA PRYMYESSES DTYMETARRT AE HYTDQDK DYNAAYTADE YNSLVKTVLL RLIEKALATL KNRLHITTID QLKKFRDYLN SDADAGEFQI FLNQEDCVIL KNL SNLASK FFNVRCVADT LEVMLEALRN NIELVQPESD AVRRIVIKMT QEIKDSSTPL YNIAMYKSDY DAIKNKNIKT LFDL YNDRL PINFLDTSAT SPVRKTSGKR SAEDDLLPTR SSKRANRPEI NVISSEDEQE DDDVEDVDYE KESKRRKLED EDFLK LKAL EFSKDIVNEK LQKIIVVTDG MKRLYEYCNC KNSLETLPSA ANYGSLLKRL NLYNLDHIEM NVNFYELLFP LTLYND NDN SDKTLSHQLV NYIFLASNYF QNCAKNFNYM RETFNVFGPF KQIDFMVMFV IKFNFLCDMR NFAKLIDELV PNKQPNM RI HSVLVMRDKI VKLAFSNLQF QTFSKKDKSR NTKHLQRLIM LMNANYNVI UniProtKB: Viral capsid associated protein |
-Macromolecule #3: AcOrf-109 peptide
| Macromolecule | Name: AcOrf-109 peptide / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Autographa californica multiple nucleopolyhedrovirus Autographa californica multiple nucleopolyhedrovirus |
| Molecular weight | Theoretical: 44.851441 KDa |
| Sequence | String: MECPFQIQVC ISDRFFAFPH NLVEPQSDVG NKLIENLIVY VPTDDDRLYI DKKQFPKFNS VLVYRHEHDV NIDSRSPKKT ASATIVYWN PLVPITEIGA GETRVFSVLL TNNLFYCNTM IVHHENPKCP IEFTYPETDM QSACSALLKN RNGQSVPPPI K SNLRPIAC ...String: MECPFQIQVC ISDRFFAFPH NLVEPQSDVG NKLIENLIVY VPTDDDRLYI DKKQFPKFNS VLVYRHEHDV NIDSRSPKKT ASATIVYWN PLVPITEIGA GETRVFSVLL TNNLFYCNTM IVHHENPKCP IEFTYPETDM QSACSALLKN RNGQSVPPPI K SNLRPIAC EIPLSHFKEL VESNDFLLCF NLETSTMVKI LSLKRIFCIF QYRKQPARYV INLPHEEIDN LYNKLNWERT RR LMKGDVP SNCATVNRSS LKYIKQAQSL LGIPDYSQTV VDFVKMFQKI IFPYQLVPNV IIKLNNFDQM VSSAPNKAEP YKK IRLFCK NDSIAISSSG IVPINMPDFS PPNTFDYSDY ANRTNINFVT QRVLTDGGFS SGITVTPVKY NYYL UniProtKB: AcOrf-109 peptide |
-Macromolecule #4: Early 49 Daa protein
| Macromolecule | Name: Early 49 Daa protein / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Autographa californica multiple nucleopolyhedrovirus Autographa californica multiple nucleopolyhedrovirus |
| Molecular weight | Theoretical: 55.480898 KDa |
| Sequence | String: MSGGGNLLTL ERDHFKYLFL TSYFDLKDNE HVPSEPMAFI RNYLNCTFDL LDDAVLMNYF NYLQSMQLKH LVGSTSTNIF KFVKPQFRF VCDRTTVDIL EFDTRMYIKP GTPVYATNLF TSNPRKMMAF LYAEFGKVFK NKIFVNINNY GCVLAGSAGF L FDDAYVDW ...String: MSGGGNLLTL ERDHFKYLFL TSYFDLKDNE HVPSEPMAFI RNYLNCTFDL LDDAVLMNYF NYLQSMQLKH LVGSTSTNIF KFVKPQFRF VCDRTTVDIL EFDTRMYIKP GTPVYATNLF TSNPRKMMAF LYAEFGKVFK NKIFVNINNY GCVLAGSAGF L FDDAYVDW NGVRMCAAPR LDNNMHPFRL YLLGEDMAKH FVDNNILPPH PSNAKTRKIN NSMFMLKNFY KGLPLFKSKY TV VNSTKIV TRKPNDIFNE IDKELNGNCP FIKFIQRDYI FDAQFPPDLL DLLNEYMTKS SIMKIITKFV IEENPAMSGE MSR EIILDR YSVDNYRKLY IKMEITNQFP VMYDHESSYI FVSKDFLQLK GTMNAFYAPK QRILSILAVN RLFGATETID FHPN LLVYR QSSPPVRLTG DVYVVDKNEK VFLVKHVFSN TVPAYLLIRG DYESSSDLKS LRDLNPWVQN TLLKLLIPDS VQ UniProtKB: Early 49 Daa protein |
-Macromolecule #5: P40
| Macromolecule | Name: P40 / type: protein_or_peptide / ID: 5 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Autographa californica multiple nucleopolyhedrovirus Autographa californica multiple nucleopolyhedrovirus |
| Molecular weight | Theoretical: 41.583594 KDa |
| Sequence | String: MSAIALYLEI NKLRLKIDEP MQLAIWPQLF PLLCDEHQSV QLNTDVLINF MMHVARKSQN TILNNNAAIA SQYAAGNADV VAAPASAQP TPRPVINLFA RANAAAPAQP SEELINMRRY RNAARKLIHH YSLNSTSSTE YKISDVVMTM IFLLRSEKYH S LFKLLETT ...String: MSAIALYLEI NKLRLKIDEP MQLAIWPQLF PLLCDEHQSV QLNTDVLINF MMHVARKSQN TILNNNAAIA SQYAAGNADV VAAPASAQP TPRPVINLFA RANAAAPAQP SEELINMRRY RNAARKLIHH YSLNSTSSTE YKISDVVMTM IFLLRSEKYH S LFKLLETT FDDYTCRPQM TQVQTDTLLD AVRSLLEMPS TTIDLTTVDI MRSSFARCFN SPIMRYAKIV LLQNVALQRD KR TTLEELL IERGEKIQML QPQQYINSGT EIPFCDDAEF LNRLLKHIDP YPLSRMYYNA ANTMFYTTME NYAVSNCKFN IED YNNIFK VMENIRKHSN KNSNDQDELN IYLGVQSSNA KRKKY UniProtKB: P40 |
-Macromolecule #6: Occlusion-derived virus envelope/capsid protein
| Macromolecule | Name: Occlusion-derived virus envelope/capsid protein / type: protein_or_peptide / ID: 6 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Autographa californica multiple nucleopolyhedrovirus Autographa californica multiple nucleopolyhedrovirus |
| Molecular weight | Theoretical: 33.568152 KDa |
| Sequence | String: MKRIKCNKVR TVTEIVNSDE KIQKTYELAE FDLKNLSSLE SYETLKIKLA LSKYMAMLST LEMTQPLLEI FRNKADTRQI AAVVFSTLA FIHNRFHPLV TNFTNKMEFV VTETNDTSIP GEPILFTENE GVLLCSVDRP SIVKMLSREF DTEALVNFEN D NCNVRIAK ...String: MKRIKCNKVR TVTEIVNSDE KIQKTYELAE FDLKNLSSLE SYETLKIKLA LSKYMAMLST LEMTQPLLEI FRNKADTRQI AAVVFSTLA FIHNRFHPLV TNFTNKMEFV VTETNDTSIP GEPILFTENE GVLLCSVDRP SIVKMLSREF DTEALVNFEN D NCNVRIAK TFGASKRKNT TRSDDYESNK QPNYDMDLSD FSITEVEATQ YLTLLLTVEH AYLHYYIFKN YGVFEYCKSL TD HSLFTNK LRSTMSTKTS NLLLSKFKFT IEDFDKINSN SVTSGFNIYN FNK UniProtKB: Occlusion-derived virus envelope/capsid protein |
-Macromolecule #7: 38K
| Macromolecule | Name: 38K / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Autographa californica multiple nucleopolyhedrovirus Autographa californica multiple nucleopolyhedrovirus |
| Molecular weight | Theoretical: 38.0705 KDa |
| Sequence | String: MASSLQSKWI CLRLNDAIIK RHVLVLSEYA DLKYLGFEKY KFFEYVIFQF CNDPQLCKII ENNYNYCMQI FKAPADDMRD IRHNIKRAF KTPVLGHMCV LSNKPPMYSF LKEWFLLPHY KVVSLKSESL TWGFPHVVVF DLDSTLITEE EQVEIRDPFV Y DSLQELHE ...String: MASSLQSKWI CLRLNDAIIK RHVLVLSEYA DLKYLGFEKY KFFEYVIFQF CNDPQLCKII ENNYNYCMQI FKAPADDMRD IRHNIKRAF KTPVLGHMCV LSNKPPMYSF LKEWFLLPHY KVVSLKSESL TWGFPHVVVF DLDSTLITEE EQVEIRDPFV Y DSLQELHE MGCVLVLWSY GSRDHVAHSM RDVDLEGYFD IIISEGSTVQ EERSDLVQNS HNAIVDYNLK KRFIENKFVF DI HNHRSDN NIPKSPKIVI KYLSDKNVNF FKSITLVDDL PTNNYAYDFY VKVKRCPTPV QDWEHYHNEI IQNIMDYEQY FIK UniProtKB: 38K |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm Bright-field microscopy / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z
Z Y
Y X
X