+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
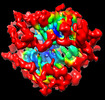
| Title | SARS-CoV-2 Delta variant spike protein | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  SARS-CoV-2 / SARS-CoV-2 /  Delta / Delta /  Spike protein / Spike protein /  VIRAL PROTEIN VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationMaturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / suppression by virus of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / entry receptor-mediated virion attachment to host cell ...Maturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / suppression by virus of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / receptor-mediated endocytosis of virus by host cell / Attachment and Entry /  membrane fusion / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / membrane fusion / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell /  receptor ligand activity / host cell surface receptor binding / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / receptor ligand activity / host cell surface receptor binding / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane /  viral envelope / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / viral envelope / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane /  membrane / identical protein binding / membrane / identical protein binding /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Severe acute respiratory syndrome coronavirus 2 Severe acute respiratory syndrome coronavirus 2 | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.1 Å cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Xu J / Cheng H / Liu N / Wang HW | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Self-assembled monolayers guided free-standing atomic-crystal/molecule superstructure Authors: Zheng L / Xu J / Wang W / Gao X / Liu N / Wang HW / Peng HL | |||||||||
| History |
|
- Structure visualization
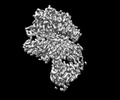
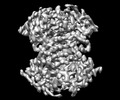
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34975.map.gz emd_34975.map.gz | 483.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34975-v30.xml emd-34975-v30.xml emd-34975.xml emd-34975.xml | 14.1 KB 14.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34975.png emd_34975.png | 83.3 KB | ||
| Filedesc metadata |  emd-34975.cif.gz emd-34975.cif.gz | 6 KB | ||
| Others |  emd_34975_half_map_1.map.gz emd_34975_half_map_1.map.gz emd_34975_half_map_2.map.gz emd_34975_half_map_2.map.gz | 474.9 MB 474.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34975 http://ftp.pdbj.org/pub/emdb/structures/EMD-34975 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34975 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34975 | HTTPS FTP |
-Related structure data
| Related structure data |  8hrjMC  8hriC  8hrkC  8hrlC  8hrmC  8hrnC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34975.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34975.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_34975_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
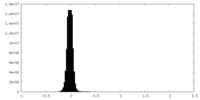
| Density Histograms |
-Half map: #1
| File | emd_34975_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
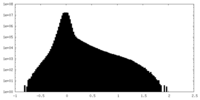
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 Delta variant spike protein
| Entire | Name: SARS-CoV-2 Delta variant spike protein |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 Delta variant spike protein
| Supramolecule | Name: SARS-CoV-2 Delta variant spike protein / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: on graphene/SAMs membranes |
|---|---|
| Source (natural) | Organism:   Severe acute respiratory syndrome coronavirus 2 Severe acute respiratory syndrome coronavirus 2 |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 Details: For the issue of poor atom inclusion, the depositor stated 'we have tried our best to fit the coordinates in the density map of spike via rigid-body dock. However, the flexibility of RBD ...Details: For the issue of poor atom inclusion, the depositor stated 'we have tried our best to fit the coordinates in the density map of spike via rigid-body dock. However, the flexibility of RBD makes it hard to fit, which seems to be general for spike proteins.' Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Severe acute respiratory syndrome coronavirus 2 Severe acute respiratory syndrome coronavirus 2 |
| Molecular weight | Theoretical: 138.842375 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MPRGPVAALL LLILHGAWSC VNLTTRTQLP PAYTNSFTRG VYYPDKVFRS SVLHSTQDLF LPFFSNVTWF HVISGTNGTK RFDNPVLPF NDGVYFASIE KSNIIRGWIF GTTLDSKTQS LLIVNNATNV VIKVCEFQFC NDPFLDHKNN KSWMESEFRV Y SSANNCTF ...String: MPRGPVAALL LLILHGAWSC VNLTTRTQLP PAYTNSFTRG VYYPDKVFRS SVLHSTQDLF LPFFSNVTWF HVISGTNGTK RFDNPVLPF NDGVYFASIE KSNIIRGWIF GTTLDSKTQS LLIVNNATNV VIKVCEFQFC NDPFLDHKNN KSWMESEFRV Y SSANNCTF EYVSQPFLMD LEGKQGNFKN LREFVFKNID GYFKIYSKHT PIIVREPEDL PQGFSALEPL VDLPIGINIT RF QTLLALH RSYLTPGDSS SGWTAGAAAY YVGYLQPRTF LLKYNENGTI TDAVDCALDP LSETKCTLKS FTVEKGIYQT SNF RVQPTE SIVRFPNITN LCPFDEVFNA TRFASVYAWN RKRISNCVAD YSVLYNLAPF FTFKCYGVSP TKLNDLCFTN VYAD SFVIR GDEVRQIAPG QTGNIADYNY KLPDDFTGCV IAWNSNKLDS KVSGNYNYLY RLFRKSNLKP FERDISTEIY QAGNK PCNG VAGFNCYFPL RSYSFRPTYG VGHQPYRVVV LSFELLHAPA TVCGPKKSTN LVKNKCVNFN FNGLKGTGVL TESNKK FLP FQQFGRDIAD TTDAVRDPQT LEILDITPCS FGGVSVITPG TNTSNQVAVL YQGVNCTEVP VAIHADQLTP TWRVYST GS NVFQTRAGCL IGAEYVNNSY ECDIPIGAGI CASYQTQTKS HGSASSVASQ SIIAYTMSLG AENSVAYSNN SIAIPTNF T ISVTTEILPV SMTKTSVDCT MYICGDSTEC SNLLLQYGSF CTQLKRALTG IAVEQDKNTQ EVFAQVKQIY KTPPIKYFG GFNFSQILPD PSKPSKRSFI EDLLFNKVTL ADAGFIKQYG DCLGDIAARD LICAQKFKGL TVLPPLLTDE MIAQYTSALL AGTITSGWT FGAGAALQIP FAMQMAYRFN GIGVTQNVLY ENQKLIANQF NSAIGKIQDS LSSTASALGK LQDVVNHNAQ A LNTLVKQL SSKFGAISSV LNDIFSRLDP PEAEVQIDRL ITGRLQSLQT YVTQQLIRAA EIRASANLAA TKMSECVLGQ SK RVDFCGK GYHLMSFPQS APHGVVFLHV TYVPAQEKNF TTAPAICHDG KAHFPREGVF VSNGTHWFVT QRNFYEPQII TTD NTFVSG NCDVVIGIVN NTVYDPLQPE LDSFKEELDK YFKNHTSPDV DLGDISGINA SVVNIQKEID RLNEVAKNLN ESLI DLQEL GKYEQLVPRG SGYIPEAPRD GQAYVRKDGE WVLLSTFLHH HHHH UniProtKB:  Spike glycoprotein Spike glycoprotein |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.25 mg/mL |
|---|---|
| Buffer | pH: 7.8 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.2 µm Bright-field microscopy / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.2 µm |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.1 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 75650 |
 Movie
Movie Controller
Controller










 Z
Z Y
Y X
X