+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Aplysia californica FaNaC in ligand bound state | |||||||||
 Map data Map data | ligand bound full map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  neuropeptide / neuropeptide /  ion channel / FMRFamide / ion channel / FMRFamide /  TRANSPORT PROTEIN TRANSPORT PROTEIN | |||||||||
| Function / homology |  Epithelial sodium channel / Epithelial sodium channel /  Amiloride-sensitive sodium channel / Amiloride-sensitive sodium channel /  sodium channel activity / sodium channel activity /  membrane / FMRFamide-gated Na+ channel membrane / FMRFamide-gated Na+ channel Function and homology information Function and homology information | |||||||||
| Biological species |   Aplysia californica (California sea hare) / synthetic construct (others) Aplysia californica (California sea hare) / synthetic construct (others) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.9 Å cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Chen QF / Liu FL / Dang Y / Feng H / Zhang Z / Ye S | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Chem Biol / Year: 2023 Journal: Nat Chem Biol / Year: 2023Title: Structure and mechanism of a neuropeptide-activated channel in the ENaC/DEG superfamily. Authors: Fenglian Liu / Yu Dang / Lu Li / Hao Feng / Jianlin Li / Haowei Wang / Xu Zhang / Zhe Zhang / Sheng Ye / Yutao Tian / Qingfeng Chen /  Abstract: Phe-Met-Arg-Phe-amide (FMRFamide)-activated sodium channels (FaNaCs) are a family of channels activated by the neuropeptide FMRFamide, and, to date, the underlying ligand gating mechanism remains ...Phe-Met-Arg-Phe-amide (FMRFamide)-activated sodium channels (FaNaCs) are a family of channels activated by the neuropeptide FMRFamide, and, to date, the underlying ligand gating mechanism remains unknown. Here we present the high-resolution cryo-electron microscopy structures of Aplysia californica FaNaC in both apo and FMRFamide-bound states. AcFaNaC forms a chalice-shaped trimer and possesses several notable features, including two FaNaC-specific insertion regions, a distinct finger domain and non-domain-swapped transmembrane helix 2 in the transmembrane domain (TMD). One FMRFamide binds to each subunit in a cleft located in the top-most region of the extracellular domain, with participation of residues from the neighboring subunit. Bound FMRFamide adopts an extended conformation. FMRFamide binds tightly to A. californica FaNaC in an N terminus-in manner, which causes collapse of the binding cleft and induces large local conformational rearrangements. Such conformational changes are propagated downward toward the TMD via the palm domain, possibly resulting in outward movement of the TMD and dilation of the ion conduction pore. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34122.map.gz emd_34122.map.gz | 59.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34122-v30.xml emd-34122-v30.xml emd-34122.xml emd-34122.xml | 19 KB 19 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34122.png emd_34122.png | 161.1 KB | ||
| Filedesc metadata |  emd-34122.cif.gz emd-34122.cif.gz | 6.2 KB | ||
| Others |  emd_34122_half_map_1.map.gz emd_34122_half_map_1.map.gz emd_34122_half_map_2.map.gz emd_34122_half_map_2.map.gz | 49.7 MB 49.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34122 http://ftp.pdbj.org/pub/emdb/structures/EMD-34122 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34122 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34122 | HTTPS FTP |
-Related structure data
| Related structure data |  7yvbMC  7yvcC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34122.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34122.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ligand bound full map | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: ligand bound half map 1
| File | emd_34122_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ligand bound half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
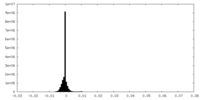
| Density Histograms |
-Half map: ligand bound half map 2
| File | emd_34122_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ligand bound half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
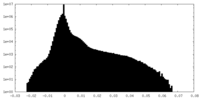
| Density Histograms |
- Sample components
Sample components
-Entire : Trimeric FMRFamide activated sodium channel from Aplysia californ...
| Entire | Name: Trimeric FMRFamide activated sodium channel from Aplysia californica (AcFaNaC) |
|---|---|
| Components |
|
-Supramolecule #1: Trimeric FMRFamide activated sodium channel from Aplysia californ...
| Supramolecule | Name: Trimeric FMRFamide activated sodium channel from Aplysia californica (AcFaNaC) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Aplysia californica (California sea hare) Aplysia californica (California sea hare) |
| Molecular weight | Theoretical: 73 KDa |
-Macromolecule #1: FMRFamide-gated Na+ channel
| Macromolecule | Name: FMRFamide-gated Na+ channel / type: protein_or_peptide / ID: 1 Details: Residues 672-679 correspond to the EXPRESSION TAG, whereas resides 660-665 correspond to the thrombin cleavage site, and resides 654-659 and 666-671 correspond to flexible linkers. Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Aplysia californica (California sea hare) Aplysia californica (California sea hare) |
| Molecular weight | Theoretical: 76.638312 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MLGRGERIKP YHFRDSSADH MKYTSVSAKS GMVPEHRYTM VRSRHHGRHH HHHSYQEYNT QRSAISLIAE LGSESNAHGL AKIVTSRDT KRKVIWALMV IIGFTAATLQ LSLLVRKYLQ FQVVELSEIK DSMPVEYPSV TICNIEPISL RKIRKAYNKN E SQNLKDWL ...String: MLGRGERIKP YHFRDSSADH MKYTSVSAKS GMVPEHRYTM VRSRHHGRHH HHHSYQEYNT QRSAISLIAE LGSESNAHGL AKIVTSRDT KRKVIWALMV IIGFTAATLQ LSLLVRKYLQ FQVVELSEIK DSMPVEYPSV TICNIEPISL RKIRKAYNKN E SQNLKDWL NFTQTFHFKD MSFMNSIRAF YENLGSDAKK ISHDLRDLLI HCRFNREECT TENFTSSFDG NYFNCFTFNG GQ LRDQLQM HATGPENGLS LIISIEKDEP LPGTYGVYNF ENNILHSAGV RVVVHAPGSM PSPVDHGFDI PPGYSSSVGL KAL LHTRLS EPYGNCTEDS LEGIQTYRNT FFACLQLCKQ RRLIRECKCK SSALPDLSVE NITFCGVIPD WKDIRRNVTG EYKM NQTIP TISLACEARV QKQLNNDRSY ETECGCYQPC SETSYLKSVS LSYWPLEFYQ LSALERFFSQ KNPTDQQHFM KIAQD FLSR LAHPQQQALA RNNSHDKDIL TTSYSLSEKE MAKEASDLIR QNLLRLNIYL EDLSVVEYRQ LPAYGLADLF ADIGGT LGL WMGISVLTIM ELMELIIRLF ALIFNAEREV PKAPVHSSNN GGGGGGDGQH NFANGDVEHE RDTHFPDLGS SDFDFRR GG GIGAESPVNV EGGSSGGLVP RGSGGSSGGH HHHHHHH UniProtKB: FMRFamide-gated Na+ channel |
-Macromolecule #2: Phe-Met-Arg-Phe-amide
| Macromolecule | Name: Phe-Met-Arg-Phe-amide / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 599.767 Da |
| Sequence | String: FMR(NFA) |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 6 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4.00 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm Bright-field microscopy / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Image recording | #0 - Image recording ID: 1 / #0 - Film or detector model: GATAN K3 (6k x 4k) / #0 - Average electron dose: 60.0 e/Å2 / #1 - Image recording ID: 2 / #1 - Film or detector model: GATAN K3 (6k x 4k) / #1 - Average electron dose: 60.0 e/Å2 / #2 - Image recording ID: 3 / #2 - Film or detector model: GATAN K3 (6k x 4k) / #2 - Average electron dose: 60.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing #1
Image processing #1
| Startup model | Type of model: NONE |
|---|---|
| Initial angle assignment | Type: OTHER |
| Final angle assignment | Type: PROJECTION MATCHING |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.9 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 267000 |
| Image processing ID | 1 |
| Image recording ID | 1 |
- Image processing #2
Image processing #2
| Startup model | Type of model: NONE |
|---|---|
| Initial angle assignment | Type: OTHER |
| Final angle assignment | Type: PROJECTION MATCHING |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.9 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 267000 |
| Image processing ID | 2 |
| Image recording ID | 2 |
- Image processing #3
Image processing #3
| Startup model | Type of model: NONE |
|---|---|
| Initial angle assignment | Type: OTHER |
| Final angle assignment | Type: PROJECTION MATCHING |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.9 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 267000 |
| Image processing ID | 3 |
| Image recording ID | 3 |
- Image processing #4
Image processing #4
| Startup model | Type of model: NONE |
|---|---|
| Initial angle assignment | Type: OTHER |
| Final angle assignment | Type: PROJECTION MATCHING |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.9 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 267000 |
| Image processing ID | 4 |
| Image recording ID | 1 |
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7yvb: |
 Movie
Movie Controller
Controller




 Z
Z Y
Y X
X