+ Open data
Open data
- Basic information
Basic information
| Entry | 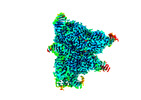 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the OgeuIscB-omega RNA-target DNA complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  metagenome (others) / synthetic construct (others) metagenome (others) / synthetic construct (others) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.55 Å cryo EM / Resolution: 2.55 Å | |||||||||
 Authors Authors | Kato K / Okazaki O / Isayama Y / Ishikawa J / Nishizawa T / Nishimasu H | |||||||||
| Funding support |  Japan, 1 items Japan, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structure of the IscB-ωRNA ribonucleoprotein complex, the likely ancestor of CRISPR-Cas9. Authors: Kazuki Kato / Sae Okazaki / Soumya Kannan / Han Altae-Tran / F Esra Demircioglu / Yukari Isayama / Junichiro Ishikawa / Masahiro Fukuda / Rhiannon K Macrae / Tomohiro Nishizawa / Kira S ...Authors: Kazuki Kato / Sae Okazaki / Soumya Kannan / Han Altae-Tran / F Esra Demircioglu / Yukari Isayama / Junichiro Ishikawa / Masahiro Fukuda / Rhiannon K Macrae / Tomohiro Nishizawa / Kira S Makarova / Eugene V Koonin / Feng Zhang / Hiroshi Nishimasu /   Abstract: Transposon-encoded IscB family proteins are RNA-guided nucleases in the OMEGA (obligate mobile element-guided activity) system, and likely ancestors of the RNA-guided nuclease Cas9 in the type II ...Transposon-encoded IscB family proteins are RNA-guided nucleases in the OMEGA (obligate mobile element-guided activity) system, and likely ancestors of the RNA-guided nuclease Cas9 in the type II CRISPR-Cas adaptive immune system. IscB associates with its cognate ωRNA to form a ribonucleoprotein complex that cleaves double-stranded DNA targets complementary to an ωRNA guide segment. Although IscB shares the RuvC and HNH endonuclease domains with Cas9, it is much smaller than Cas9, mainly due to the lack of the α-helical nucleic-acid recognition lobe. Here, we report the cryo-electron microscopy structure of an IscB protein from the human gut metagenome (OgeuIscB) in complex with its cognate ωRNA and a target DNA, at 2.6-Å resolution. This high-resolution structure reveals the detailed architecture of the IscB-ωRNA ribonucleoprotein complex, and shows how the small IscB protein assembles with the ωRNA and mediates RNA-guided DNA cleavage. The large ωRNA scaffold structurally and functionally compensates for the recognition lobe of Cas9, and participates in the recognition of the guide RNA-target DNA heteroduplex. These findings provide insights into the mechanism of the programmable DNA cleavage by the IscB-ωRNA complex and the evolution of the type II CRISPR-Cas9 effector complexes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33198.map.gz emd_33198.map.gz | 118 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33198-v30.xml emd-33198-v30.xml emd-33198.xml emd-33198.xml | 17.6 KB 17.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33198.png emd_33198.png | 63.1 KB | ||
| Others |  emd_33198_half_map_1.map.gz emd_33198_half_map_1.map.gz emd_33198_half_map_2.map.gz emd_33198_half_map_2.map.gz | 116 MB 116 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33198 http://ftp.pdbj.org/pub/emdb/structures/EMD-33198 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33198 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33198 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33198.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33198.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_33198_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_33198_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Structure of the IscB-omegaRNA ribonucleoprotein complex
| Entire | Name: Structure of the IscB-omegaRNA ribonucleoprotein complex |
|---|---|
| Components |
|
-Supramolecule #1: Structure of the IscB-omegaRNA ribonucleoprotein complex
| Supramolecule | Name: Structure of the IscB-omegaRNA ribonucleoprotein complex type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: #4 |
|---|
-Supramolecule #2: IscB-omegaRNA
| Supramolecule | Name: IscB-omegaRNA / type: complex / Chimera: Yes / ID: 2 / Parent: 1 / Macromolecule list: #4 |
|---|---|
| Source (natural) | Organism:  metagenome (others) metagenome (others) |
-Supramolecule #3: DNA
| Supramolecule | Name: DNA / type: complex / Chimera: Yes / ID: 3 / Parent: 1 / Macromolecule list: #3 |
|---|
-Macromolecule #1: RNA (228-MER)
| Macromolecule | Name: RNA (228-MER) / type: rna / ID: 1 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  metagenome (others) metagenome (others) |
| Molecular weight | Theoretical: 73.640742 KDa |
| Sequence | String: UGUGAGCGGA UAACAAUUCC CCGGCUCUUC CAACUUUAUG GUUGCGACCG UAGGUUGAAA GAGCACAGGC UGAGACAUUC GUAAGGCCG AAAGACCGGA CGCACCCUGG GAUUUCCCCA GUCCCCGGAA CUGCAUAGCG GAUGCCAGUU GAUGGAGCAA U CUAUCAGA ...String: UGUGAGCGGA UAACAAUUCC CCGGCUCUUC CAACUUUAUG GUUGCGACCG UAGGUUGAAA GAGCACAGGC UGAGACAUUC GUAAGGCCG AAAGACCGGA CGCACCCUGG GAUUUCCCCA GUCCCCGGAA CUGCAUAGCG GAUGCCAGUU GAUGGAGCAA U CUAUCAGA UAAGCCAGGG GGAACAAUCA CCUCUCUGUA UCAGAGAGAG UUUUACAAAA GGAGGAACGG |
-Macromolecule #2: DNA (49-MER)
| Macromolecule | Name: DNA (49-MER) / type: dna / ID: 2 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 15.092702 KDa |
| Sequence | String: (DG)(DA)(DA)(DT)(DG)(DG)(DT)(DT)(DT)(DT) (DC)(DT)(DT)(DC)(DG)(DG)(DG)(DG)(DA)(DA) (DT)(DT)(DG)(DT)(DT)(DA)(DT)(DC)(DC) (DG)(DC)(DT)(DC)(DA)(DC)(DA)(DA)(DT)(DT) (DC) (DC)(DT)(DT)(DA)(DG)(DA)(DA)(DA) (DA) |
-Macromolecule #3: DNA (5'-D(P*GP*AP*AP*GP*AP*AP*AP*AP*CP*CP*AP*T)-3')
| Macromolecule | Name: DNA (5'-D(P*GP*AP*AP*GP*AP*AP*AP*AP*CP*CP*AP*T)-3') / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 4.281832 KDa |
| Sequence | String: (DG)(DA)(DA)(DG)(DA)(DA)(DA)(DA)(DC)(DC) (DA)(DT)(DT)(DC) |
-Macromolecule #4: OgeuIscB
| Macromolecule | Name: OgeuIscB / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  metagenome (others) metagenome (others) |
| Molecular weight | Theoretical: 56.81968 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MMAVVYVISK SGKPLMPTTR CGHVRILLKE GKARVVERKP FTIQLTYESA EETQPLVLGI DPGRTNIGMS VVTESGESVF NAQIETRNK DVPKLMKDRK QYRMAHRRLK RRCKRRRRAK AAGTAFEEGE KQRLLPGCFK PITCKSIRNK EARFNNRKRP V GWLTPTAN ...String: MMAVVYVISK SGKPLMPTTR CGHVRILLKE GKARVVERKP FTIQLTYESA EETQPLVLGI DPGRTNIGMS VVTESGESVF NAQIETRNK DVPKLMKDRK QYRMAHRRLK RRCKRRRRAK AAGTAFEEGE KQRLLPGCFK PITCKSIRNK EARFNNRKRP V GWLTPTAN HLLVTHLNVV KKVQKILPVA KVVLELNRFS FMAMNNPKVQ RWQYQRGPLY GKGSVEEAVS MQQDGHCLFC KH GIDHYHH VVPRRKNGSE TLENRVGLCE EHHRLVHTDK EWEANLASKK SGMNKKYHAL SVLNQIIPYL ADQLADMFPG NFC VTSGQD TYLFREEHGI PKDHYLDAYC IACSALTDAK KVSSPKGRPY MVHQFRRHDR QACHKANLNR SYYMGGKLVA TNRH KAMDQ KTDSLEEYRA AHSAADVSKL TVKHPSAQYK DMSRIMPGSI LVSGEGKLFT LSRSEGRNKG QVNYFVSTEG IKYWA RKCQ YLRNNGGLQI YV |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #6: LAURYL DIMETHYLAMINE-N-OXIDE
| Macromolecule | Name: LAURYL DIMETHYLAMINE-N-OXIDE / type: ligand / ID: 6 / Number of copies: 1 / Formula: LDA |
|---|---|
| Molecular weight | Theoretical: 229.402 Da |
| Chemical component information |  ChemComp-LDA: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DIFFRACTION / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 47.9 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
|---|---|
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.55 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 792405 |
 Movie
Movie Controller
Controller







 Z
Z Y
Y X
X

















