+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
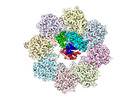
| Title | An apo TRiC map | |||||||||
 Map data Map data | A sharpened map of apo TRiC | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.11 Å cryo EM / Resolution: 3.11 Å | |||||||||
 Authors Authors | Park JS / Roh SH | |||||||||
| Funding support |  Korea, Republic Of, 1 items Korea, Republic Of, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2022 Journal: Cell / Year: 2022Title: Structural visualization of the tubulin folding pathway directed by human chaperonin TRiC/CCT. Authors: Daniel Gestaut / Yanyan Zhao / Junsun Park / Boxue Ma / Alexander Leitner / Miranda Collier / Grigore Pintilie / Soung-Hun Roh / Wah Chiu / Judith Frydman /    Abstract: The ATP-dependent ring-shaped chaperonin TRiC/CCT is essential for cellular proteostasis. To uncover why some eukaryotic proteins can only fold with TRiC assistance, we reconstituted the folding of ...The ATP-dependent ring-shaped chaperonin TRiC/CCT is essential for cellular proteostasis. To uncover why some eukaryotic proteins can only fold with TRiC assistance, we reconstituted the folding of β-tubulin using human prefoldin and TRiC. We find unstructured β-tubulin is delivered by prefoldin to the open TRiC chamber followed by ATP-dependent chamber closure. Cryo-EM resolves four near-atomic-resolution structures containing progressively folded β-tubulin intermediates within the closed TRiC chamber, culminating in native tubulin. This substrate folding pathway appears closely guided by site-specific interactions with conserved regions in the TRiC chamber. Initial electrostatic interactions between the TRiC interior wall and both the folded tubulin N domain and its C-terminal E-hook tail establish the native substrate topology, thus enabling C-domain folding. Intrinsically disordered CCT C termini within the chamber promote subsequent folding of tubulin's core and middle domains and GTP-binding. Thus, TRiC's chamber provides chemical and topological directives that shape the folding landscape of its obligate substrates. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32822.map.gz emd_32822.map.gz | 117.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32822-v30.xml emd-32822-v30.xml emd-32822.xml emd-32822.xml | 25 KB 25 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_32822_fsc.xml emd_32822_fsc.xml | 11.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_32822.png emd_32822.png | 111.9 KB | ||
| Others |  emd_32822_half_map_1.map.gz emd_32822_half_map_1.map.gz emd_32822_half_map_2.map.gz emd_32822_half_map_2.map.gz | 115.6 MB 115.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32822 http://ftp.pdbj.org/pub/emdb/structures/EMD-32822 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32822 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32822 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_32822.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32822.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | A sharpened map of apo TRiC | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: A half map of apo TRiC
| File | emd_32822_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | A half map of apo TRiC | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: A half map of apo TRiC
| File | emd_32822_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | A half map of apo TRiC | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Apo TRiC/CCT
| Entire | Name: Apo TRiC/CCT |
|---|---|
| Components |
|
-Supramolecule #1: Apo TRiC/CCT
| Supramolecule | Name: Apo TRiC/CCT / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1 MDa |
-Macromolecule #1: CCT subunit 1
| Macromolecule | Name: CCT subunit 1 / type: other / ID: 1 / Classification: other |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEGPLSVFGD RSTGETIRSQ NVMAAASIAN IVKSSLGPVG LDKMLVDDIG DVTITNDGAT ILKLLEVEH PAAKVLCELA DLQDKEVGDG TTSVVIIAAE LLKNADELVK QKIHPTSVIS G YRLACKEA VRYINENLIV NTDELGRDCL INAAKTSMSS KIIGINGDFF ...String: MEGPLSVFGD RSTGETIRSQ NVMAAASIAN IVKSSLGPVG LDKMLVDDIG DVTITNDGAT ILKLLEVEH PAAKVLCELA DLQDKEVGDG TTSVVIIAAE LLKNADELVK QKIHPTSVIS G YRLACKEA VRYINENLIV NTDELGRDCL INAAKTSMSS KIIGINGDFF ANMVVDAVLA IK YTDIRGQ PRYPVNSVNI LKAHGRSQME SMLISGYALN CVVGSQGMPK RIVNAKIACL DFS LQKTKM KLGVQVVITD PEKLDQIRQR ESDITKERIQ KILATGANVI LTTGGIDDMC LKYF VEAGA MAVRRVLKRD LKRIAKASGA TILSTLANLE GEETFEAAML GQAEEVVQER ICDDE LILI KNTKARTSAS IILRGANDFM CDEMERSLHD ALCVVKRVLE SKSVVPGGGA VEAALS IYL ENYATSMGSR EQLAIAEFAR SLLVIPNTLA VNAAQDSTDL VAKLRAFHNE AQVNPER KN LKWIGLDLSN GKPRDNKQAG VFEPTIVKVK SLKFATEAAI TILRIDDLIK LHPESKDD K HGSYEDAVHS GALND |
| Recombinant expression | Organism:   Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
-Macromolecule #2: CCT subunit 2
| Macromolecule | Name: CCT subunit 2 / type: other / ID: 2 / Classification: other |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MASLSLAPVN IFKAGADEER AETARLTSFI GAIAIGDLVK STLGPKGMDK ILLSSGRDAS LMVTNDGAT ILKNIGVDNP AAKVLVDMSR VQDDEVGDGT TSVTVLAAEL LREAESLIAK K IHPQTIIA GWREATKAAR EALLSSAVDH GSDEVKFRQD LMNIAGTTLS ...String: MASLSLAPVN IFKAGADEER AETARLTSFI GAIAIGDLVK STLGPKGMDK ILLSSGRDAS LMVTNDGAT ILKNIGVDNP AAKVLVDMSR VQDDEVGDGT TSVTVLAAEL LREAESLIAK K IHPQTIIA GWREATKAAR EALLSSAVDH GSDEVKFRQD LMNIAGTTLS SKLLTHHKDH FT KLAVEAV LRLKGSGNLE AIHIIKKLGG SLADSYLDEG FLLDKKIGVN QPKRIENAKI LIA NTGMDT DKIKIFGSRV RVDSTAKVAE IEHAEKEKMK EKVERILKHG INCFINRQLI YNYP EQLFG AAGVMAIEHA DFAGVERLAL VTGGEIASTF DHPELVKLGS CKLIEEVMIG EDKLI HFSG VALGEACTIV LRGATQQILD EAERSLHDAL CVLAQTVKDS RTVYGGGCSE MLMAHA VTQ LANRTPGKEA VAMESYAKAL RMLPTIIADN AGYDSADLVA QLRAAHSEGN TTAGLDM RE GTIGDMAILG ITESFQVKRQ VLLSAAEAAE VILRVDNIIK AAPRKRVPDH HPC |
| Recombinant expression | Organism:   Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
-Macromolecule #3: CCT subunit 3
| Macromolecule | Name: CCT subunit 3 / type: other / ID: 3 / Classification: other |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MMGHRPVLVL SQNTKRESGR KVQSGNINAA KTIADIIRTC LGPKSMMKML LDPMGGIVMT NDGNAILRE IQVQHPAAKS MIEISRTQDE EVGDGTTSVI ILAGEMLSVA EHFLEQQMHP T VVISAYRK ALDDMISTLK KISIPVDISD SDMMLNIINS SITTKAISRW ...String: MMGHRPVLVL SQNTKRESGR KVQSGNINAA KTIADIIRTC LGPKSMMKML LDPMGGIVMT NDGNAILRE IQVQHPAAKS MIEISRTQDE EVGDGTTSVI ILAGEMLSVA EHFLEQQMHP T VVISAYRK ALDDMISTLK KISIPVDISD SDMMLNIINS SITTKAISRW SSLACNIALD AV KMVQFEE NGRKEIDIKK YARVEKIPGG IIEDSCVLRG VMINKDVTHP RMRRYIKNPR IVL LDSSLE YKKGESQTDI EITREEDFTR ILQMEEEYIQ QLCEDIIQLK PDVVITEKGI SDLA QHYLM RANITAIRRV RKTDNNRIAR ACGARIVSRP EELREDDVGT GAGLLEIKKI GDEYF TFIT DCKDPKACTI LLRGASKEIL SEVERNLQDA MQVCRNVLLD PQLVPGGGAS EMAVAH ALT EKSKAMTGVE QWPYRAVAQA LEVIPRTLIQ NCGASTIRLL TSLRAKHTQE NCETWGV NG ETGTLVDMKE LGIWEPLAVK LQTYKTAVET AVLLLRIDDI VSGHKKKGDD QSRQGGAP D AGQE |
| Recombinant expression | Organism:   Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
-Macromolecule #4: CCT subunit 4
| Macromolecule | Name: CCT subunit 4 / type: other / ID: 4 / Classification: other |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MPENVAPRSG ATAGAAGGRG KGAYQDRDKP AQIRFSNISA AKAVADAIRT SLGPKGMDKM IQDGKGDVT ITNDGATILK QMQVLHPAAR MLVELSKAQD IEAGDGTTSV VIIAGSLLDS C TKLLQKGI HPTIISESFQ KALEKGIEIL TDMSRPVELS DRETLLNSAT ...String: MPENVAPRSG ATAGAAGGRG KGAYQDRDKP AQIRFSNISA AKAVADAIRT SLGPKGMDKM IQDGKGDVT ITNDGATILK QMQVLHPAAR MLVELSKAQD IEAGDGTTSV VIIAGSLLDS C TKLLQKGI HPTIISESFQ KALEKGIEIL TDMSRPVELS DRETLLNSAT TSLNSKVVSQ YS SLLSPMS VNAVMKVIDP ATATSVDLRD IKIVKKLGGT IDDCELVEGL VLTQKVSNSG ITR VEKAKI GLIQFCLSAP KTDMDNQIVV SDYAQMDRVL REERAYILNL VKQIKKTGCN VLLI QKSIL RDALSDLALH FLNKMKIMVI KDIEREDIEF ICKTIGTKPV AHIDQFTADM LGSAE LAEE VNLNGSGKLL KITGCASPGK TVTIVVRGSN KLVIEEAERS IHDALCVIRC LVKKRA LIA GGGAPEIELA LRLTEYSRTL SGMESYCVRA FADAMEVIPS TLAENAGLNP ISTVTEL RN RHAQGEKTAG INVRKGGISN ILEELVVQPL LVSVSALTLA TETVRSILKI DDVVNTR |
| Recombinant expression | Organism:   Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
-Macromolecule #5: CCT subunit 5
| Macromolecule | Name: CCT subunit 5 / type: other / ID: 5 / Classification: other |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MASMGTLAFD EYGRPFLIIK DQDRKSRLMG LEALKSHIMA AKAVANTMRT SLGPNGLDKM MVDKDGDVT VTNDGATILS MMDVDHQIAK LMVELSKSQD DEIGDGTTGV VVLAGALLEE A EQLLDRGI HPIRIADGYE QAARVAIEHL DKISDSVLVD IKDTEPLIQT ...String: MASMGTLAFD EYGRPFLIIK DQDRKSRLMG LEALKSHIMA AKAVANTMRT SLGPNGLDKM MVDKDGDVT VTNDGATILS MMDVDHQIAK LMVELSKSQD DEIGDGTTGV VVLAGALLEE A EQLLDRGI HPIRIADGYE QAARVAIEHL DKISDSVLVD IKDTEPLIQT AKTTLGSKVV NS CHRQMAE IAVNAVLTVA DMERRDVDFE LIKVEGKVGG RLEDTKLIKG VIVDKDFSHP QMP KKVEDA KIAILTCPFE PPKPKTKHKL DVTSVEDYKA LQKYEKEKFE EMIQQIKETG ANLA ICQWG FDDEANHLLL QNNLPAVRWV GGPEIELIAI ATGGRIVPRF SELTAEKLGF AGLVQ EISF GTTKDKMLVI EQCKNSRAVT IFIRGGNKMI IEEAKRSLHD ALCVIRNLIR DNRVVY GGG AAEISCALAV SQEADKCPTL EQYAMRAFAD ALEVIPMALS ENSGMNPIQT MTEVRAR QV KEMNPALGID CLHKGTNDMK QQHVIETLIG KKQQISLATQ MVRMILKIDD IRKPGESEE |
| Recombinant expression | Organism:   Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
-Macromolecule #6: CCT subunit 6
| Macromolecule | Name: CCT subunit 6 / type: other / ID: 6 / Classification: other |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAAVKTLNPK AEVARAQAAL AVNISAARGL QDVLRTNLGP KGTMKMLVSG AGDIKLTKDG NVLLHEMQI QHPTASLIAK VATAQDDITG DGTTSNVLII GELLKQADLY ISEGLHPRII T EGFEAAKE KALQFLEEVK VSREMDRETL IDVARTSLRT KVHAELADVL ...String: MAAVKTLNPK AEVARAQAAL AVNISAARGL QDVLRTNLGP KGTMKMLVSG AGDIKLTKDG NVLLHEMQI QHPTASLIAK VATAQDDITG DGTTSNVLII GELLKQADLY ISEGLHPRII T EGFEAAKE KALQFLEEVK VSREMDRETL IDVARTSLRT KVHAELADVL TEAVVDSILA IK KQDEPID LFMIEIMEMK HKSETDTSLI RGLVLDHGAR HPDMKKRVED AYILTCNVSL EYE KTEVNS GFFYKSAEER EKLVKAERKF IEDRVKKIIE LKRKVCGDSD KGFVVINQKG IDPF SLDAL SKEGIVALRR AKRRNMERLT LACGGVALNS FDDLSPDCLG HAGLVYEYTL GEEKF TFIE KCNNPRSVTL LIKGPNKHTL TQIKDAVRDG LRAVKNAIDD GCVVPGAGAV EVAMAE ALI KHKPSVKGRA QLGVQAFADA LLIIPKVLAQ NSGFDLQETL VKIQAEHSES GQLVGVD LN TGEPMVAAEV GVWDNYCVKK QLLHSCTVIA TNILLVDEIM RAGMSFLKG |
| Recombinant expression | Organism:   Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
-Macromolecule #7: CCT subunit 7
| Macromolecule | Name: CCT subunit 7 / type: other / ID: 7 / Classification: other |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MMPTPVILLK EGTDSSQGIP QLVSNISACQ VIAEAVRTTL GPRGMDKLIV DGRGKATISN DGATILKLL DVVHPAAKTL VDIAKSQDAE VGDGTTSVTL LAAEFLKQVK PYVEEGLHPQ I IIRAFRTA TQLAVNKIKE IAVTVKKADK VEQRKLLEKC AMTALSSKLI ...String: MMPTPVILLK EGTDSSQGIP QLVSNISACQ VIAEAVRTTL GPRGMDKLIV DGRGKATISN DGATILKLL DVVHPAAKTL VDIAKSQDAE VGDGTTSVTL LAAEFLKQVK PYVEEGLHPQ I IIRAFRTA TQLAVNKIKE IAVTVKKADK VEQRKLLEKC AMTALSSKLI SQQKAFFAKM VV DAVMMLD DLLQLKMIGI KKVQGGALED SQLVAGVAFK KTFSYAGFEM QPKKYHNPKI ALL NVELEL KAEKDNAEIR VHTVEDYQAI VDAEWNILYD KLEKIHHSGA KVVLSKLPIG DVAT QYFAD RDMFCAGRVP EEDLKRTMMA CGGSIQTSVN ALSADVLGRC QVFEETQIGG ERYNF FTGC PKAKTCTFIL RGGAEQFMEE TERSLHDAIM IVRRAIKNDS VVAGGGAIEM ELSKYL RDY SRTIPGKQQL LIGAYAKALE IIPRQLCDNA GFDATNILNK LRARHAQGGS HHHHHHG SG TWYGVDINNE DIADNFEAFV WEPAMVRINA LTAASEAACL IVSVDETIKN PRSTVDAP T AAGRGRGRGR PH |
| Recombinant expression | Organism:   Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
-Macromolecule #8: CCT subunit 8
| Macromolecule | Name: CCT subunit 8 / type: other / ID: 8 / Classification: other |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MALHVPKAPG FAQMLKEGAK HFSGLEEAVY RNIQACKELA QTTRTAYGPN GMNKMVINHL EKLFVTNDA ATILRELEVQ HPAAKMIVMA SHMQEQEVGD GTNFVLVFAG ALLELAEELL R IGLSVSEV IEGYEIACRK AHEILPNLVC CSAKNLRDID EVSSLLRTSI ...String: MALHVPKAPG FAQMLKEGAK HFSGLEEAVY RNIQACKELA QTTRTAYGPN GMNKMVINHL EKLFVTNDA ATILRELEVQ HPAAKMIVMA SHMQEQEVGD GTNFVLVFAG ALLELAEELL R IGLSVSEV IEGYEIACRK AHEILPNLVC CSAKNLRDID EVSSLLRTSI MSKQYGNEVF LA KLIAQAC VSIFPDSGHF NVDNIRVCKI LGSGISSSSV LHGMVFKKET EGDVTSVKDA KIA VYSCPF DGMITETKGT VLIKTAEELM NFSKGEENLM DAQVKAIADT GANVVVTGGK VADM ALHYA NKYNIMLVRL NSKWDLRRLC KTVGATALPR LTPPVLEEMG HCDSVYLSEV GDTQV VVFK HEKEDGAIST IVLRGSTDNL MDDIERAVDD GVNTFKVLTR DKRLVPGGGA TEIELA KQI TSYGETCPGL EQYAIKKFAE AFEAIPRALA ENSGVKANEV ISKLYAVHQE GNKNVGL DI EAEVPAVKDM LEAGILDTYL GKYWAIKLAT NAAVTVLRVD QIIMAKPAGG PKPPSGKK D WDDDQN |
| Recombinant expression | Organism:   Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | 3D array |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Component:
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Support film - Material: CARBON / Support film - topology: HOLEY | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 0.5 µm Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number real images: 6084 / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|
 Movie
Movie Controller
Controller













 Z
Z Y
Y X
X


















