+ Open data
Open data
- Basic information
Basic information
| Entry | 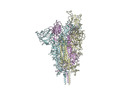 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
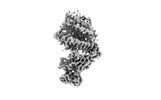
| Title | Neutral Omicron Spike Trimer | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationMaturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / suppression by virus of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / entry receptor-mediated virion attachment to host cell ...Maturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / suppression by virus of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / receptor-mediated endocytosis of virus by host cell / Attachment and Entry /  membrane fusion / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / membrane fusion / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell /  receptor ligand activity / host cell surface receptor binding / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / receptor ligand activity / host cell surface receptor binding / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane /  viral envelope / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / viral envelope / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane /  membrane / identical protein binding / membrane / identical protein binding /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Severe acute respiratory syndrome coronavirus 2 Severe acute respiratory syndrome coronavirus 2 | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.4 Å cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Cui Z / Wang X | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2022 Journal: Cell / Year: 2022Title: Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron. Authors: Zhen Cui / Pan Liu / Nan Wang / Lei Wang / Kaiyue Fan / Qianhui Zhu / Kang Wang / Ruihong Chen / Rui Feng / Zijing Jia / Minnan Yang / Ge Xu / Boling Zhu / Wangjun Fu / Tianming Chu / Leilei ...Authors: Zhen Cui / Pan Liu / Nan Wang / Lei Wang / Kaiyue Fan / Qianhui Zhu / Kang Wang / Ruihong Chen / Rui Feng / Zijing Jia / Minnan Yang / Ge Xu / Boling Zhu / Wangjun Fu / Tianming Chu / Leilei Feng / Yide Wang / Xinran Pei / Peng Yang / Xiaoliang Sunney Xie / Lei Cao / Yunlong Cao / Xiangxi Wang /  Abstract: The SARS-CoV-2 Omicron variant with increased fitness is spreading rapidly worldwide. Analysis of cryo-EM structures of the spike (S) from Omicron reveals amino acid substitutions forging ...The SARS-CoV-2 Omicron variant with increased fitness is spreading rapidly worldwide. Analysis of cryo-EM structures of the spike (S) from Omicron reveals amino acid substitutions forging interactions that stably maintain an active conformation for receptor recognition. The relatively more compact domain organization confers improved stability and enhances attachment but compromises the efficiency of the viral fusion step. Alterations in local conformation, charge, and hydrophobic microenvironments underpin the modulation of the epitopes such that they are not recognized by most NTD- and RBD-antibodies, facilitating viral immune escape. Structure of the Omicron S bound with human ACE2, together with the analysis of sequence conservation in ACE2 binding region of 25 sarbecovirus members, as well as heatmaps of the immunogenic sites and their corresponding mutational frequencies, sheds light on conserved and structurally restrained regions that can be used for the development of broad-spectrum vaccines and therapeutics. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32478.map.gz emd_32478.map.gz | 13.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32478-v30.xml emd-32478-v30.xml emd-32478.xml emd-32478.xml | 11.1 KB 11.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_32478.png emd_32478.png | 44.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32478 http://ftp.pdbj.org/pub/emdb/structures/EMD-32478 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32478 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32478 | HTTPS FTP |
-Related structure data
| Related structure data |  7wg6MC  7wg7C  7wg8C  7wg9C  7wgbC  7wgcC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32478.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32478.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : neutral Omicron Spike
| Entire | Name: neutral Omicron Spike |
|---|---|
| Components |
|
-Supramolecule #1: neutral Omicron Spike
| Supramolecule | Name: neutral Omicron Spike / type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Severe acute respiratory syndrome coronavirus 2 Severe acute respiratory syndrome coronavirus 2 |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Severe acute respiratory syndrome coronavirus 2 Severe acute respiratory syndrome coronavirus 2 |
| Molecular weight | Theoretical: 127.821125 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QCVNLTTRTQ LPPAYTNSFT RGVYYPDKVF RSSVLHSTQD LFLPFFSNVT WFHVIHVSGT NGTKRFDNPV LPFNDGVYFA SIEKSNIIR GWIFGTTLDS KTQSLLIVNN ATNVVIKVCE FQFCNDPFLD HKNNKSWMES EFRVYSSANN CTFEYVSQPF L MDLEGKQG ...String: QCVNLTTRTQ LPPAYTNSFT RGVYYPDKVF RSSVLHSTQD LFLPFFSNVT WFHVIHVSGT NGTKRFDNPV LPFNDGVYFA SIEKSNIIR GWIFGTTLDS KTQSLLIVNN ATNVVIKVCE FQFCNDPFLD HKNNKSWMES EFRVYSSANN CTFEYVSQPF L MDLEGKQG NFKNLREFVF KNIDGYFKIY SKHTPIIVRE PEDLPQGFSA LEPLVDLPIG INITRFQTLL ALHRSYLTPG DS SSGWTAG AAAYYVGYLQ PRTFLLKYNE NGTITDAVDC ALDPLSETKC TLKSFTVEKG IYQTSNFRVQ PTESIVRFPN ITN LCPFDE VFNATRFASV YAWNRKRISN CVADYSVLYN LAPFFTFKCY GVSPTKLNDL CFTNVYADSF VIRGDEVRQI APGQ TGKIA DYNYKLPDDF TGCVIAWNSN KLDSKVSGNY NYLYRLFRKS NLKPFERDIS TEIYQAGNKP CNGVAGFNCY FPLRS YSFR PTYGVGHQPY RVVVLSFELL HAPATVCGPK KSTNLVKNKC VNFNFNGLKG TGVLTESNKK FLPFQQFGRD IADTTD AVR DPQTLEILDI TPCSFGGVSV ITPGTNTSNQ VAVLYQGVNC TEVPVAIHAD QLTPTWRVYS TGSNVFQTRA GCLIGAE YV NNSYECDIPI GAGICASYQT QTNSPRRARS VASQSIIAYT MSLGAENSVA YSNNSIAIPT NFTISVTTEI LPVSMTKT S VDCTMYICGD STECSNLLLQ YGSFCTQLNR ALTGIAVEQD KNTQEVFAQV KQIYKTPPIK YFGGFNFSQI LPDPSKPSK RSFIEDLLFN KVTLADAGFI KQYGDCLGDI AARDLICAQK FKGLTVLPPL LTDEMIAQYT SALLAGTITS GWTFGAGAAL QIPFAMQMA YRFNGIGVTQ NVLYENQKLI ANQFNSAIGK IQDSLSSTAS ALGKLQDVVN HNAQALNTLV KQLSSKFGAI S SVLNDIFS RLDKVEAEVQ IDRLITGRLQ SLQTYVTQQL IRAAEIRASA NLAATKMSEC VLGQSKRVDF CGKGYHLMSF PQ SAPHGVV FLHVTYVPAQ EKNFTTAPAI CHDGKAHFPR EGVFVSNGTH WFVTQRNFYE PQIITTDNTF VSGNCDVVIG IVN NTVYDP LQPELDSFKE ELDKYFKNHT SP |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 24 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
|---|---|
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.4 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 221744 |
 Movie
Movie Controller
Controller