[English] 日本語
 Yorodumi
Yorodumi- EMDB-32193: STRUCTURE OF PHOTOSYNTHETIC LH1-RC SUPER-COMPLEX OF RHODOBACTER S... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | STRUCTURE OF PHOTOSYNTHETIC LH1-RC SUPER-COMPLEX OF RHODOBACTER SPHAEROIDES LACKING PROTEIN-U | |||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationorganelle inner membrane / plasma membrane-derived chromatophore membrane / plasma membrane light-harvesting complex /  bacteriochlorophyll binding / photosynthetic electron transport in photosystem II / bacteriochlorophyll binding / photosynthetic electron transport in photosystem II /  photosynthesis, light reaction / electron transporter, transferring electrons within the cyclic electron transport pathway of photosynthesis activity / membrane => GO:0016020 / photosynthesis, light reaction / electron transporter, transferring electrons within the cyclic electron transport pathway of photosynthesis activity / membrane => GO:0016020 /  metal ion binding / metal ion binding /  plasma membrane plasma membraneSimilarity search - Function | |||||||||||||||||||||
| Biological species |  Rhodobacter sphaeroides f. sp. denitrificans (bacteria) Rhodobacter sphaeroides f. sp. denitrificans (bacteria) | |||||||||||||||||||||
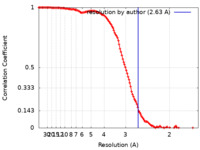
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.63 Å cryo EM / Resolution: 2.63 Å | |||||||||||||||||||||
 Authors Authors | Tani K / Kanno R / Kawamura S / Kikuchi R / Nagashima KVP / Hall M / Takahashi A / Yu L-J / Kimura Y / Madigan MT ...Tani K / Kanno R / Kawamura S / Kikuchi R / Nagashima KVP / Hall M / Takahashi A / Yu L-J / Kimura Y / Madigan MT / Mizoguchi A / Humbel BM / Wang-Otomo Z-Y | |||||||||||||||||||||
| Funding support |  Japan, 6 items Japan, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Asymmetric structure of the native Rhodobacter sphaeroides dimeric LH1-RC complex. Authors: Kazutoshi Tani / Ryo Kanno / Riku Kikuchi / Saki Kawamura / Kenji V P Nagashima / Malgorzata Hall / Ai Takahashi / Long-Jiang Yu / Yukihiro Kimura / Michael T Madigan / Akira Mizoguchi / ...Authors: Kazutoshi Tani / Ryo Kanno / Riku Kikuchi / Saki Kawamura / Kenji V P Nagashima / Malgorzata Hall / Ai Takahashi / Long-Jiang Yu / Yukihiro Kimura / Michael T Madigan / Akira Mizoguchi / Bruno M Humbel / Zheng-Yu Wang-Otomo /    Abstract: Rhodobacter sphaeroides is a model organism in bacterial photosynthesis, and its light-harvesting-reaction center (LH1-RC) complex contains both dimeric and monomeric forms. Here we present cryo-EM ...Rhodobacter sphaeroides is a model organism in bacterial photosynthesis, and its light-harvesting-reaction center (LH1-RC) complex contains both dimeric and monomeric forms. Here we present cryo-EM structures of the native LH1-RC dimer and an LH1-RC monomer lacking protein-U (ΔU). The native dimer reveals several asymmetric features including the arrangement of its two monomeric components, the structural integrity of protein-U, the overall organization of LH1, and rigidities of the proteins and pigments. PufX plays a critical role in connecting the two monomers in a dimer, with one PufX interacting at its N-terminus with another PufX and an LH1 β-polypeptide in the other monomer. One protein-U was only partially resolved in the dimeric structure, signaling different degrees of disorder in the two monomers. The ΔU LH1-RC monomer was half-moon-shaped and contained 11 α- and 10 β-polypeptides, indicating a critical role for protein-U in controlling the number of αβ-subunits required for dimer assembly and stabilization. These features are discussed in relation to membrane topology and an assembly model proposed for the native dimeric complex. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32193.map.gz emd_32193.map.gz | 167 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32193-v30.xml emd-32193-v30.xml emd-32193.xml emd-32193.xml | 23.1 KB 23.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_32193_fsc.xml emd_32193_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_32193.png emd_32193.png | 135.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32193 http://ftp.pdbj.org/pub/emdb/structures/EMD-32193 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32193 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32193 | HTTPS FTP |
-Related structure data
| Related structure data |  7vy3MC  7vy2C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32193.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32193.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
+Entire : Photosynthetic LH1-RC complex from the purple phototrophic bacter...
+Supramolecule #1: Photosynthetic LH1-RC complex from the purple phototrophic bacter...
+Supramolecule #2: Rhodobacter sphaeroides monomeric LH1-RC Complex lacking protein-U
+Macromolecule #1: Photosynthetic reaction center L subunit
+Macromolecule #2: Reaction center protein M chain
+Macromolecule #3: Photosynthetic reaction center subunit H
+Macromolecule #4: Antenna pigment protein alpha chain
+Macromolecule #5: Antenna pigment protein beta chain
+Macromolecule #6: PufX
+Macromolecule #7: BACTERIOCHLOROPHYLL A
+Macromolecule #8: BACTERIOPHEOPHYTIN A
+Macromolecule #9: UBIQUINONE-10
+Macromolecule #10: (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-...
+Macromolecule #11: LAURYL DIMETHYLAMINE-N-OXIDE
+Macromolecule #12: DODECYL-BETA-D-MALTOSIDE
+Macromolecule #13: FE (III) ION
+Macromolecule #14: SPHEROIDENE
+Macromolecule #15: CARDIOLIPIN
+Macromolecule #16: PHOSPHATIDYLETHANOLAMINE
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.0 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 277 K / Instrument: LEICA EM GP |
| Details | This sample was monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy Bright-field microscopy |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Average exposure time: 1.275 sec. / Average electron dose: 42.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller