[English] 日本語
 Yorodumi
Yorodumi- EMDB-29930: T. cruzi topoisomerase II alpha bound to dsDNA and the covalent i... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | T. cruzi topoisomerase II alpha bound to dsDNA and the covalent inhibitor CT1 | |||||||||
 Map data Map data | Final refined and sharpened map from NU refinement. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Topoisomerase / Topoisomerase /  DNA binding protein / Topoisomerase inhibitor / DNA binding protein / Topoisomerase inhibitor /  ISOMERASE ISOMERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationDNA topoisomerase type II (double strand cut, ATP-hydrolyzing) activity /  DNA topoisomerase (ATP-hydrolysing) / DNA topological change / DNA topoisomerase (ATP-hydrolysing) / DNA topological change /  DNA binding / DNA binding /  ATP binding / ATP binding /  metal ion binding metal ion bindingSimilarity search - Function | |||||||||
| Biological species |   Trypanosoma cruzi (eukaryote) / Trypanosoma cruzi (eukaryote) /   Escherichia coli (E. coli) / Escherichia coli (E. coli) /   Trypanosoma cruzi strain CL Brener (eukaryote) Trypanosoma cruzi strain CL Brener (eukaryote) | |||||||||
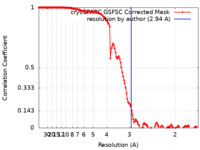
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.94 Å cryo EM / Resolution: 2.94 Å | |||||||||
 Authors Authors | Schenk A / Deniston C / Noeske J | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2023 Journal: Science / Year: 2023Title: Cyanotriazoles are selective topoisomerase II poisons that rapidly cure trypanosome infections. Authors: Srinivasa P S Rao / Matthew K Gould / Jonas Noeske / Manuel Saldivia / Rajiv S Jumani / Pearly S Ng / Olivier René / Yen-Liang Chen / Marcel Kaiser / Ryan Ritchie / Amanda Fortes Francisco ...Authors: Srinivasa P S Rao / Matthew K Gould / Jonas Noeske / Manuel Saldivia / Rajiv S Jumani / Pearly S Ng / Olivier René / Yen-Liang Chen / Marcel Kaiser / Ryan Ritchie / Amanda Fortes Francisco / Nila Johnson / Debjani Patra / Harry Cheung / Colin Deniston / Andreas D Schenk / Wilian A Cortopassi / Remo S Schmidt / Natalie Wiedemar / Bryanna Thomas / Rima Palkar / Nahdiyah A Ghafar / Vanessa Manoharan / Catherine Luu / Jonathan E Gable / Kah Fei Wan / Elmarie Myburgh / Jeremy C Mottram / Whitney Barnes / John Walker / Charles Wartchow / Natasha Aziz / Colin Osborne / Juergen Wagner / Christopher Sarko / John M Kelly / Ujjini H Manjunatha / Pascal Mäser / Jan Jiricek / Suresh B Lakshminarayana / Michael P Barrett / Thierry T Diagana /     Abstract: Millions who live in Latin America and sub-Saharan Africa are at risk of trypanosomatid infections, which cause Chagas disease and human African trypanosomiasis (HAT). Improved HAT treatments are ...Millions who live in Latin America and sub-Saharan Africa are at risk of trypanosomatid infections, which cause Chagas disease and human African trypanosomiasis (HAT). Improved HAT treatments are available, but Chagas disease therapies rely on two nitroheterocycles, which suffer from lengthy drug regimens and safety concerns that cause frequent treatment discontinuation. We performed phenotypic screening against trypanosomes and identified a class of cyanotriazoles (CTs) with potent trypanocidal activity both in vitro and in mouse models of Chagas disease and HAT. Cryo-electron microscopy approaches confirmed that CT compounds acted through selective, irreversible inhibition of trypanosomal topoisomerase II by stabilizing double-stranded DNA:enzyme cleavage complexes. These findings suggest a potential approach toward successful therapeutics for the treatment of Chagas disease. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29930.map.gz emd_29930.map.gz | 203.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29930-v30.xml emd-29930-v30.xml emd-29930.xml emd-29930.xml | 24.9 KB 24.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29930_fsc.xml emd_29930_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_29930.png emd_29930.png | 143.5 KB | ||
| Masks |  emd_29930_msk_1.map emd_29930_msk_1.map | 216 MB |  Mask map Mask map | |
| Others |  emd_29930_half_map_1.map.gz emd_29930_half_map_1.map.gz emd_29930_half_map_2.map.gz emd_29930_half_map_2.map.gz | 200.3 MB 200.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29930 http://ftp.pdbj.org/pub/emdb/structures/EMD-29930 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29930 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29930 | HTTPS FTP |
-Related structure data
| Related structure data |  8gccMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29930.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29930.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final refined and sharpened map from NU refinement. | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.845 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_29930_msk_1.map emd_29930_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Final half map from NU refinement.
| File | emd_29930_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final half map from NU refinement. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Final half map from NU refinement.
| File | emd_29930_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final half map from NU refinement. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Topoisomerase II alpha bound to dsDNA and inhibitor CT1
| Entire | Name: Topoisomerase II alpha bound to dsDNA and inhibitor CT1 |
|---|---|
| Components |
|
-Supramolecule #1: Topoisomerase II alpha bound to dsDNA and inhibitor CT1
| Supramolecule | Name: Topoisomerase II alpha bound to dsDNA and inhibitor CT1 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Trypanosoma cruzi (eukaryote) Trypanosoma cruzi (eukaryote) |
-Supramolecule #2: Topoisomerase II alpha
| Supramolecule | Name: Topoisomerase II alpha / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Trypanosoma cruzi (eukaryote) Trypanosoma cruzi (eukaryote) |
-Supramolecule #3: dsDNA
| Supramolecule | Name: dsDNA / type: complex / ID: 3 / Parent: 2 / Macromolecule list: #2 Details: DNA sequence: GG GAT AAC AAT GAG CTC ATT GTT ATC CC |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
-Macromolecule #1: DNA topoisomerase 2
| Macromolecule | Name: DNA topoisomerase 2 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Trypanosoma cruzi strain CL Brener (eukaryote) Trypanosoma cruzi strain CL Brener (eukaryote) |
| Molecular weight | Theoretical: 89.322438 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: GRNADRKQIL GIPKLDDANE AGGKYSHRCT LILTEGDSAK ALCTAGLAVK DRDYFGVFPL RGKPLNVRDA TLKKVMACAE FQAVSKIMG LDIRQKYSGV ERLRYGHLMI MSDQDHDGSH IKGLIINMIH HYWPDLIKTP GFLQQFITPI VKARKKGRSD G DDRAISFF ...String: GRNADRKQIL GIPKLDDANE AGGKYSHRCT LILTEGDSAK ALCTAGLAVK DRDYFGVFPL RGKPLNVRDA TLKKVMACAE FQAVSKIMG LDIRQKYSGV ERLRYGHLMI MSDQDHDGSH IKGLIINMIH HYWPDLIKTP GFLQQFITPI VKARKKGRSD G DDRAISFF SMPDYFEWKN AIGDGIRNYE IRYYKGLGTS GAKEGREYFE NIDRHRLDFV HEDATDDARI VMAFAKDKVE ER KHWITQF KANTNVNESM NYNVRTVRYS EFVDKELILF SVADCERSIP SVIDGLKPGQ RKIIFSSFKR RLTRSIKVVQ LAG YVSEHA AYHHGEQSLV QTIVGLAQNF VGSNNVPLLQ QDGQFGTRLQ GGKDHAAGRY IFTRLTNIAR YIYHPSDDFV VDYK DDDGL SVEPFYYVPV IPMVLVNGTS GIGTGFATNI PNYSPLEVID NLMRLLRGEE VQPMKPWYFG FAGTIEEKEK GKFVS TGCA NVRPDGVVQI TELPIGTWTQ GYKKFLEELR EKEVVVQYRE HNTDVTVDFE VFLHPEVLHH WVAQGCVEER LQLREY IHA TNIIAFDREG QITKYRDAEA VLKEFYLVRL EYYAKRRDFL IGDLRSVASK LENMVRFVTE VVDGRLIVTR RRKKELL EE LRQRGYAPFP LQQKKKVSST TIQQGEEEGA ADATHATAED VFLVLQPAVD EGGDEDNQET PEMRRAARDY DYLLGMRL W NLTAEMIARL QSQLQKARDE LAALEKRTPK DLWAEDLNQL RPRIENLFEE RAKEIASI UniProtKB:  DNA topoisomerase 2 DNA topoisomerase 2 |
-Macromolecule #2: DNA (28-MER)
| Macromolecule | Name: DNA (28-MER) / type: dna / ID: 2 / Number of copies: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Molecular weight | Theoretical: 8.604566 KDa |
| Sequence | String: (DG)(DG)(DG)(DA)(DT)(DA)(DA)(DC)(DA)(DA) (DT)(DG)(DA)(DG)(DC)(DT)(DC)(DA)(DT)(DT) (DG)(DT)(DT)(DA)(DT)(DC)(DC)(DC) |
-Macromolecule #3: 2-{3-[(Z)-iminomethyl]-1H-1,2,4-triazol-1-yl}-1-{(3M)-3-[2-(trifl...
| Macromolecule | Name: 2-{3-[(Z)-iminomethyl]-1H-1,2,4-triazol-1-yl}-1-{(3M)-3-[2-(trifluoromethyl)phenyl]-6H-pyrrolo[3,4-b]pyridin-6-yl}ethan-1-one type: ligand / ID: 3 / Number of copies: 2 / Formula: YWX |
|---|---|
| Molecular weight | Theoretical: 398.341 Da |
| Chemical component information |  ChemComp-YWX: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.3 Component:
| |||||||||||||||
| Grid | Model: UltrAuFoil / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 90 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.026000000000000002 kPa | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 0.01 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 75000 Bright-field microscopy / Cs: 0.01 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Image recording | #0 - Image recording ID: 1 / #0 - Film or detector model: FEI FALCON IV (4k x 4k) / #0 - Digitization - Dimensions - Width: 4096 pixel / #0 - Digitization - Dimensions - Height: 4096 pixel / #0 - Number grids imaged: 1 / #0 - Number real images: 3014 / #0 - Average exposure time: 2.8 sec. / #0 - Average electron dose: 50.0 e/Å2 #0 - Details: Micrographs collected at 30 degrees tilt using a Falcon 4i camera. #1 - Image recording ID: 2 / #1 - Film or detector model: FEI FALCON IV (4k x 4k) / #1 - Digitization - Dimensions - Width: 4096 pixel / #1 - Digitization - Dimensions - Height: 4096 pixel / #1 - Number grids imaged: 1 / #1 - Number real images: 2220 / #1 - Average exposure time: 2.98 sec. / #1 - Average electron dose: 50.0 e/Å2 #1 - Details: Micrographs collected at 30 degrees tilt using a Falcon 4 camera. |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z
Z Y
Y X
X