[English] 日本語
 Yorodumi
Yorodumi- EMDB-28963: LH2-LH3 antenna in parallel configuration embedded in a nanodisc -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | LH2-LH3 antenna in parallel configuration embedded in a nanodisc | ||||||||||||||||||
 Map data Map data | parallel LHII-LHIII dimer in nanodisk | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | antenna /  membrane protein / membrane protein /  nanodisc / nanodisc /  bacteriochlorophyll / bacteriochlorophyll /  PHOTOSYNTHESIS PHOTOSYNTHESIS | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationorganelle inner membrane / plasma membrane light-harvesting complex /  bacteriochlorophyll binding / bacteriochlorophyll binding /  photosynthesis, light reaction / electron transporter, transferring electrons within the cyclic electron transport pathway of photosynthesis activity / photosynthesis, light reaction / electron transporter, transferring electrons within the cyclic electron transport pathway of photosynthesis activity /  metal ion binding / metal ion binding /  plasma membrane plasma membraneSimilarity search - Function | ||||||||||||||||||
| Biological species |  Magnetospirillum molischianum (magnetotactic) Magnetospirillum molischianum (magnetotactic) | ||||||||||||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 11.3 Å cryo EM / Resolution: 11.3 Å | ||||||||||||||||||
 Authors Authors | Toporik H / Harris D / Schlau-Cohen GS / Mazor Y | ||||||||||||||||||
| Funding support |  United States, 5 items United States, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: Elucidating interprotein energy transfer dynamics within the antenna network from purple bacteria. Authors: Dihao Wang / Olivia C Fiebig / Dvir Harris / Hila Toporik / Yi Ji / Chern Chuang / Muath Nairat / Ashley L Tong / John I Ogren / Stephanie M Hart / Jianshu Cao / James N Sturgis / Yuval ...Authors: Dihao Wang / Olivia C Fiebig / Dvir Harris / Hila Toporik / Yi Ji / Chern Chuang / Muath Nairat / Ashley L Tong / John I Ogren / Stephanie M Hart / Jianshu Cao / James N Sturgis / Yuval Mazor / Gabriela S Schlau-Cohen /   Abstract: In photosynthesis, absorbed light energy transfers through a network of antenna proteins with near-unity quantum efficiency to reach the reaction center, which initiates the downstream biochemical ...In photosynthesis, absorbed light energy transfers through a network of antenna proteins with near-unity quantum efficiency to reach the reaction center, which initiates the downstream biochemical reactions. While the energy transfer dynamics within individual antenna proteins have been extensively studied over the past decades, the dynamics between the proteins are poorly understood due to the heterogeneous organization of the network. Previously reported timescales averaged over such heterogeneity, obscuring individual interprotein energy transfer steps. Here, we isolated and interrogated interprotein energy transfer by embedding two variants of the primary antenna protein from purple bacteria, light-harvesting complex 2 (LH2), together into a near-native membrane disc, known as a nanodisc. We integrated ultrafast transient absorption spectroscopy, quantum dynamics simulations, and cryogenic electron microscopy to determine interprotein energy transfer timescales. By varying the diameter of the nanodiscs, we replicated a range of distances between the proteins. The closest distance possible between neighboring LH2, which is the most common in native membranes, is 25 Å and resulted in a timescale of 5.7 ps. Larger distances of 28 to 31 Å resulted in timescales of 10 to 14 ps. Corresponding simulations showed that the fast energy transfer steps between closely spaced LH2 increase transport distances by ∼15%. Overall, our results introduce a framework for well-controlled studies of interprotein energy transfer dynamics and suggest that protein pairs serve as the primary pathway for the efficient transport of solar energy. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28963.map.gz emd_28963.map.gz | 9.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28963-v30.xml emd-28963-v30.xml emd-28963.xml emd-28963.xml | 18.3 KB 18.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_28963.png emd_28963.png | 26.6 KB | ||
| Others |  emd_28963_half_map_1.map.gz emd_28963_half_map_1.map.gz emd_28963_half_map_2.map.gz emd_28963_half_map_2.map.gz | 9.8 MB 9.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28963 http://ftp.pdbj.org/pub/emdb/structures/EMD-28963 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28963 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28963 | HTTPS FTP |
-Related structure data
| Related structure data |  8fbbMC  7tuwC  7tv3C  8fb9C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28963.map.gz / Format: CCP4 / Size: 12.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28963.map.gz / Format: CCP4 / Size: 12.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | parallel LHII-LHIII dimer in nanodisk | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.75 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_28963_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
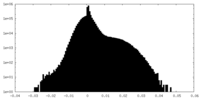
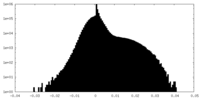
| Density Histograms |
-Half map: #2
| File | emd_28963_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
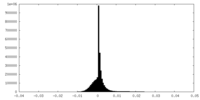
| Density Histograms |
- Sample components
Sample components
-Entire : LHII-LHIII complex in nanodisk
| Entire | Name: LHII-LHIII complex in nanodisk |
|---|---|
| Components |
|
-Supramolecule #1: LHII-LHIII complex in nanodisk
| Supramolecule | Name: LHII-LHIII complex in nanodisk / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Magnetospirillum molischianum (magnetotactic) Magnetospirillum molischianum (magnetotactic) |
| Molecular weight | Theoretical: 200 KDa |
-Macromolecule #1: Light-harvesting protein B800-820 alpha chain
| Macromolecule | Name: Light-harvesting protein B800-820 alpha chain / type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Magnetospirillum molischianum (magnetotactic) Magnetospirillum molischianum (magnetotactic) |
| Molecular weight | Theoretical: 6.08812 KDa |
| Sequence | String: SNPKDDYKIW LVINPSTWLP VIWIVALLTA IAVHSFVLSV PGYNFLASAA AKTAAK UniProtKB: Light-harvesting protein B800-820 alpha chain |
-Macromolecule #2: Light-harvesting protein B800-820 beta chain
| Macromolecule | Name: Light-harvesting protein B800-820 beta chain / type: protein_or_peptide / ID: 2 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Magnetospirillum molischianum (magnetotactic) Magnetospirillum molischianum (magnetotactic) |
| Molecular weight | Theoretical: 5.120873 KDa |
| Sequence | String: AERSLSGLTE EEAVAVHDQF KTTFSAFILL AAVAHVLVWI WKPWF UniProtKB: Light-harvesting protein B800-820 beta chain |
-Macromolecule #3: Light-harvesting protein B-800/850 alpha chain
| Macromolecule | Name: Light-harvesting protein B-800/850 alpha chain / type: protein_or_peptide / ID: 3 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Magnetospirillum molischianum (magnetotactic) Magnetospirillum molischianum (magnetotactic) |
| Molecular weight | Theoretical: 5.94498 KDa |
| Sequence | String: SNPKDDYKIW LVINPSTWLP VIWIVATVVA IAVHAAVLAA PGFNWIALGA AKSAAK UniProtKB: Light-harvesting protein B-800/850 alpha chain |
-Macromolecule #4: Light-harvesting protein B-800/850 beta 1 chain
| Macromolecule | Name: Light-harvesting protein B-800/850 beta 1 chain / type: protein_or_peptide / ID: 4 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Magnetospirillum molischianum (magnetotactic) Magnetospirillum molischianum (magnetotactic) |
| Molecular weight | Theoretical: 5.120873 KDa |
| Sequence | String: AERSLSGLTE EEAIAVHDQF KTTFSAFIIL AAVAHVLVWV WKPWF UniProtKB: Light-harvesting protein B-800/850 beta 1 chain |
-Macromolecule #5: BACTERIOCHLOROPHYLL A
| Macromolecule | Name: BACTERIOCHLOROPHYLL A / type: ligand / ID: 5 / Number of copies: 48 / Formula: BCL |
|---|---|
| Molecular weight | Theoretical: 911.504 Da |
| Chemical component information |  ChemComp-BCL: |
-Macromolecule #6: LYCOPENE
| Macromolecule | Name: LYCOPENE / type: ligand / ID: 6 / Number of copies: 16 / Formula: LYC |
|---|---|
| Molecular weight | Theoretical: 536.873 Da |
| Chemical component information |  ChemComp-LYC: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.75 µm Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.75 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 75.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION (ver. 4.0) |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 11.3 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 11133 |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-8fbb: |
 Movie
Movie Controller
Controller






 Z
Z Y
Y X
X