+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | MicroED structure of triclinic lysozyme | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationLactose synthesis /  Antimicrobial peptides / Neutrophil degranulation / Antimicrobial peptides / Neutrophil degranulation /  beta-N-acetylglucosaminidase activity / cell wall macromolecule catabolic process / beta-N-acetylglucosaminidase activity / cell wall macromolecule catabolic process /  lysozyme / lysozyme /  lysozyme activity / killing of cells of another organism / defense response to Gram-negative bacterium / defense response to Gram-positive bacterium ...Lactose synthesis / lysozyme activity / killing of cells of another organism / defense response to Gram-negative bacterium / defense response to Gram-positive bacterium ...Lactose synthesis /  Antimicrobial peptides / Neutrophil degranulation / Antimicrobial peptides / Neutrophil degranulation /  beta-N-acetylglucosaminidase activity / cell wall macromolecule catabolic process / beta-N-acetylglucosaminidase activity / cell wall macromolecule catabolic process /  lysozyme / lysozyme /  lysozyme activity / killing of cells of another organism / defense response to Gram-negative bacterium / defense response to Gram-positive bacterium / defense response to bacterium / lysozyme activity / killing of cells of another organism / defense response to Gram-negative bacterium / defense response to Gram-positive bacterium / defense response to bacterium /  endoplasmic reticulum / endoplasmic reticulum /  extracellular space / identical protein binding / extracellular space / identical protein binding /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Gallus gallus (chicken) / Gallus gallus (chicken) /   chicken (chicken) chicken (chicken) | |||||||||
| Method |  electron crystallography / electron crystallography /  cryo EM cryo EM | |||||||||
 Authors Authors | Clabbers MTB / Martynowycz MW / Hattne J / Gonen T | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: J Struct Biol X / Year: 2022 Journal: J Struct Biol X / Year: 2022Title: Hydrogens and hydrogen-bond networks in macromolecular MicroED data. Authors: Max T B Clabbers / Michael W Martynowycz / Johan Hattne / Tamir Gonen /  Abstract: Microcrystal electron diffraction (MicroED) is a powerful technique utilizing electron cryo-microscopy (cryo-EM) for protein structure determination of crystalline samples too small for X-ray ...Microcrystal electron diffraction (MicroED) is a powerful technique utilizing electron cryo-microscopy (cryo-EM) for protein structure determination of crystalline samples too small for X-ray crystallography. Electrons interact with the electrostatic potential of the sample, which means that the scattered electrons carry information about the charged state of atoms and provide relatively stronger contrast for visualizing hydrogen atoms. Accurately identifying the positions of hydrogen atoms, and by extension the hydrogen bonding networks, is of importance for understanding protein structure and function, in particular for drug discovery. However, identification of individual hydrogen atom positions typically requires atomic resolution data, and has thus far remained elusive for macromolecular MicroED. Recently, we presented the structure of triclinic hen egg-white lysozyme at 0.87 Å resolution. The corresponding data were recorded under low exposure conditions using an electron-counting detector from thin crystalline lamellae. Here, using these subatomic resolution MicroED data, we identified over a third of all hydrogen atom positions based on strong difference peaks, and directly visualize hydrogen bonding interactions and the charged states of residues. Furthermore, we find that the hydrogen bond lengths are more accurately described by the inter-nuclei distances than the centers of mass of the corresponding electron clouds. We anticipate that MicroED, coupled with ongoing advances in data collection and refinement, can open further avenues for structural biology by uncovering the hydrogen atoms and hydrogen bonding interactions underlying protein structure and function. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26596.map.gz emd_26596.map.gz | 32 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26596-v30.xml emd-26596-v30.xml emd-26596.xml emd-26596.xml | 13.4 KB 13.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_26596.png emd_26596.png | 505.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26596 http://ftp.pdbj.org/pub/emdb/structures/EMD-26596 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26596 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26596 | HTTPS FTP |
-Related structure data
| Related structure data |  7ulyMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26596.map.gz / Format: CCP4 / Size: 63.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26596.map.gz / Format: CCP4 / Size: 63.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X: 0.21136 Å / Y: 0.21333 Å / Z: 0.20631 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Lysozyme
| Entire | Name: Lysozyme |
|---|---|
| Components |
|
-Supramolecule #1: Lysozyme
| Supramolecule | Name: Lysozyme / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Gallus gallus (chicken) Gallus gallus (chicken) |
| Molecular weight | Theoretical: 14.4 KDa |
-Macromolecule #1: Lysozyme C
| Macromolecule | Name: Lysozyme C / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number:  lysozyme lysozyme |
|---|---|
| Source (natural) | Organism:   chicken (chicken) chicken (chicken) |
| Molecular weight | Theoretical: 14.33116 KDa |
| Sequence | String: KVFGRCELAA AMKRHGLDNY RGYSLGNWVC AAKFESNFNT QATNRNTDGS TDYGILQINS RWWCNDGRTP GSRNLCNIPC SALLSSDIT ASVNCAKKIV SDGNGMNAWV AWRNRCKGTD VQAWIRGCRL |
-Macromolecule #2: NITRATE ION
| Macromolecule | Name: NITRATE ION / type: ligand / ID: 2 / Number of copies: 4 / Formula: NO3 |
|---|---|
| Molecular weight | Theoretical: 62.005 Da |
| Chemical component information | 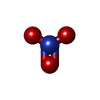 ChemComp-NO3: |
-Macromolecule #3: water
| Macromolecule | Name: water / type: ligand / ID: 3 / Number of copies: 112 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  electron crystallography electron crystallography |
| Aggregation state | 3D array |
- Sample preparation
Sample preparation
| Concentration | 10 mg/mL |
|---|---|
| Buffer | pH: 4.5 |
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 10.0 nm |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: LEICA PLUNGER |
| Details | Protein crystal lamellae |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: DIFFRACTION / Cs: 2.7 mm / Camera length: 1536 mm / Cs: 2.7 mm / Camera length: 1536 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Temperature | Min: 77.0 K / Max: 90.0 K |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Digitization - Dimensions - Width: 2048 pixel / Digitization - Dimensions - Height: 2048 pixel / Number grids imaged: 1 / Number real images: 1 / Number diffraction images: 840 / Average exposure time: 0.5 sec. / Average electron dose: 0.001 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Crystallography statistics | Number intensities measured: 569407 / Number structure factors: 64974 / Fourier space coverage: 87.58 / R sym: 0.073 / R merge: 0.236 / Overall phase error: 30 / Overall phase residual: 0 / Phase error rejection criteria: None / High resolution: 0.87 Å / Shell - Shell ID: 1 / Shell - High resolution: 0.87 Å / Shell - Low resolution: 0.9 Å / Shell - Number structure factors: 2783 / Shell - Phase residual: 30 / Shell - Fourier space coverage: 37.64 / Shell - Multiplicity: 2.1 |
|---|---|
| Final reconstruction | Resolution method: DIFFRACTION PATTERN/LAYERLINES / Software - Name: REFMAC (ver. 5.8.0267) |
| Merging software list | Software - Name: AIMLESS |
| Details | Binned by 2 |
-Atomic model buiding 1
| Refinement | Space: RECIPROCAL / Protocol: AB INITIO MODEL / Overall B value: 11.981 / Target criteria: Maximum likelihood |
|---|---|
| Output model |  PDB-7uly: |
 Movie
Movie Controller
Controller









