[English] 日本語
 Yorodumi
Yorodumi- EMDB-18699: Human H3 nucleosome assembled on alpha-satellite DNA (Class1: mos... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
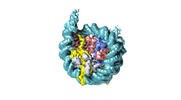
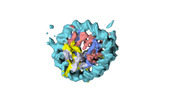
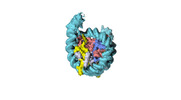
| Title | Human H3 nucleosome assembled on alpha-satellite DNA (Class1: most wrapped DNA) | |||||||||
 Map data Map data | Human H3 nucleosome assembled on aplha-satellite DNA | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Nucleosome / Nucleosome /  canonical / H3 / canonical / H3 /  Histones / Histones /  Human / Human /  DNA / DNA /  DNA BINDING PROTEIN DNA BINDING PROTEIN | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.73 Å cryo EM / Resolution: 2.73 Å | |||||||||
 Authors Authors | Ali-Ahmad A / Sekulic N | |||||||||
| Funding support |  Norway, 2 items Norway, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: CENP-A and CENP-B collaborate to create an open centromeric chromatin state. Authors: Harsh Nagpal / Ahmad Ali-Ahmad / Yasuhiro Hirano / Wei Cai / Mario Halic / Tatsuo Fukagawa / Nikolina Sekulić / Beat Fierz /     Abstract: Centromeres are epigenetically defined via the presence of the histone H3 variant CENP-A. Contacting CENP-A nucleosomes, the constitutive centromere associated network (CCAN) and the kinetochore ...Centromeres are epigenetically defined via the presence of the histone H3 variant CENP-A. Contacting CENP-A nucleosomes, the constitutive centromere associated network (CCAN) and the kinetochore assemble, connecting the centromere to spindle microtubules during cell division. The DNA-binding centromeric protein CENP-B is involved in maintaining centromere stability and, together with CENP-A, shapes the centromeric chromatin state. The nanoscale organization of centromeric chromatin is not well understood. Here, we use single-molecule fluorescence and cryoelectron microscopy (cryoEM) to show that CENP-A incorporation establishes a dynamic and open chromatin state. The increased dynamics of CENP-A chromatin create an opening for CENP-B DNA access. In turn, bound CENP-B further opens the chromatin fiber structure and induces nucleosomal DNA unwrapping. Finally, removal of CENP-A increases CENP-B mobility in cells. Together, our studies show that the two centromere-specific proteins collaborate to reshape chromatin structure, enabling the binding of centromeric factors and establishing a centromeric chromatin state. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18699.map.gz emd_18699.map.gz | 88.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18699-v30.xml emd-18699-v30.xml emd-18699.xml emd-18699.xml | 17.2 KB 17.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18699_fsc.xml emd_18699_fsc.xml | 11.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_18699.png emd_18699.png | 68.7 KB | ||
| Masks |  emd_18699_msk_1.map emd_18699_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-18699.cif.gz emd-18699.cif.gz | 5.2 KB | ||
| Others |  emd_18699_half_map_1.map.gz emd_18699_half_map_1.map.gz emd_18699_half_map_2.map.gz emd_18699_half_map_2.map.gz | 165.1 MB 165.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18699 http://ftp.pdbj.org/pub/emdb/structures/EMD-18699 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18699 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18699 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_18699.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18699.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human H3 nucleosome assembled on aplha-satellite DNA | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.704 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_18699_msk_1.map emd_18699_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_18699_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_18699_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human H3 nucleosome assembled on alpha-satellite DNA
| Entire | Name: Human H3 nucleosome assembled on alpha-satellite DNA |
|---|---|
| Components |
|
-Supramolecule #1: Human H3 nucleosome assembled on alpha-satellite DNA
| Supramolecule | Name: Human H3 nucleosome assembled on alpha-satellite DNA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: H3 histone
| Macromolecule | Name: H3 histone / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: GSMARTKQTA RKSTGGKAPR KQLATKAARK SAPSTGGVKK PHRYRPGTVA LREIRRYQKS TELLIRKLPF QRLVREIAQD FKTDLRFQSA AIGALQEASE AYLVGLFEDT NLCAIHAKRV TIMPKDIQLA RRIRGERA |
-Macromolecule #2: H4 histone
| Macromolecule | Name: H4 histone / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: GSMSGRGKGG KGLGKGGAKR HRKVLRDNIQ GITKPAIRRL ARRGGVKRIS GLIYEETRGV LKVFLENVIR DAVTYTEHAK RKTVTAMDVV YALKRQGRTL YGFGG |
-Macromolecule #3: H2A histone
| Macromolecule | Name: H2A histone / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: SGMKETAAAK FERQHMDSPD LGTLEVLFQG PLGMSGRGKQ GGKARAKAKS RSSRAGLQFP VGRVHRLLRK GNYAERVGAG APVYMAAVLE YLTAEILELA GNAARDNKKT RIIPRHLQLA IRNDEELNKL LGKVTIAQGG VLPNIQAVLL PKKTESHHKA KGK |
-Macromolecule #4: H2B histone
| Macromolecule | Name: H2B histone / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MPEPAKSAPA PKKGSKKAVT KAQKKDGKKR KRSRKESYSV YVYKVLKQVH PDTGISSKAM GIMNSFVNDI FERIAGEASR LAHYNKRSTI TSREIQTAVR LLLPGELAKH AVSEGTKAVT KYTSSK |
-Macromolecule #5: alpha-satellite DNA
| Macromolecule | Name: alpha-satellite DNA / type: dna / ID: 5 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GGAGGATTTC GTTGGAAACG GGATCAACTT CCCATAACTG AACGGAAGCA AACTCAGAAC ATTCTTTGTG ATGTTTGTAT TCAACTCACA GAGTTGAACC TTCCTTTGAT AGTTCAGGTT TGCAACACCC TTGTAGTAGA ATCTGCAAGT GTATATTTTG ACCACTTTGG A |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.1 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 6 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.2 µm Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.2 µm |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z
Z Y
Y X
X


























