[English] 日本語
 Yorodumi
Yorodumi- EMDB-17436: Photorhabdus luminescens Makes caterpillars floppy (Mcf) toxin wi... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Photorhabdus luminescens Makes caterpillars floppy (Mcf) toxin with the C-terminal deletion | |||||||||
 Map data Map data | The primary map that was used for model building | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Bacterial toxin / Bacterial toxin /  TOXIN TOXIN | |||||||||
| Function / homology | TcdA/TcdB toxin, pore forming domain / TcdA/TcdB pore forming domain /  Toxin protein Toxin protein Function and homology information Function and homology information | |||||||||
| Biological species |   Photorhabdus luminescens (bacteria) Photorhabdus luminescens (bacteria) | |||||||||
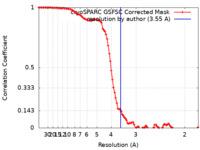
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.55 Å cryo EM / Resolution: 3.55 Å | |||||||||
 Authors Authors | Belyy A / Heilen P / Hofnagel O / Raunser S | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structure and activation mechanism of the Makes caterpillars floppy 1 toxin. Authors: Alexander Belyy / Philipp Heilen / Philine Hagel / Oliver Hofnagel / Stefan Raunser /  Abstract: The bacterial Makes caterpillars floppy 1 (Mcf1) toxin promotes apoptosis in insects, leading to loss of body turgor and death. The molecular mechanism underlying Mcf1 intoxication is poorly ...The bacterial Makes caterpillars floppy 1 (Mcf1) toxin promotes apoptosis in insects, leading to loss of body turgor and death. The molecular mechanism underlying Mcf1 intoxication is poorly understood. Here, we present the cryo-EM structure of Mcf1 from Photorhabdus luminescens, revealing a seahorse-like shape with a head and tail. While the three head domains contain two effectors, as well as an activator-binding domain (ABD) and an autoprotease, the tail consists of two putative translocation and three putative receptor-binding domains. Rearrangement of the tail moves the C-terminus away from the ABD and allows binding of the host cell ADP-ribosylation factor 3, inducing conformational changes that position the cleavage site closer to the protease. This distinct activation mechanism that is based on a hook-loop interaction results in three autocleavage reactions and the release of two toxic effectors. Unexpectedly, the BH3-like domain containing ABD is not an active effector. Our findings allow us to understand key steps of Mcf1 intoxication at the molecular level. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17436.map.gz emd_17436.map.gz | 51.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17436-v30.xml emd-17436-v30.xml emd-17436.xml emd-17436.xml | 19.5 KB 19.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17436_fsc.xml emd_17436_fsc.xml | 9.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_17436.png emd_17436.png | 98 KB | ||
| Masks |  emd_17436_msk_1.map emd_17436_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17436.cif.gz emd-17436.cif.gz | 7.3 KB | ||
| Others |  emd_17436_additional_1.map.gz emd_17436_additional_1.map.gz emd_17436_half_map_1.map.gz emd_17436_half_map_1.map.gz emd_17436_half_map_2.map.gz emd_17436_half_map_2.map.gz | 51.7 MB 95.6 MB 95.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17436 http://ftp.pdbj.org/pub/emdb/structures/EMD-17436 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17436 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17436 | HTTPS FTP |
-Related structure data
| Related structure data |  8p51MC  8p50C  8p52C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17436.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17436.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The primary map that was used for model building | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.9 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17436_msk_1.map emd_17436_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Local refinement map that was used to build...
| File | emd_17436_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local refinement map that was used to build the N-terminal effector region | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: First half map
| File | emd_17436_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | First half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Second half map
| File | emd_17436_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Second half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Photorhabdus luminescens Makes caterpillars floppy (Mcf) toxin wi...
| Entire | Name: Photorhabdus luminescens Makes caterpillars floppy (Mcf) toxin with the C-terminal deletion |
|---|---|
| Components |
|
-Supramolecule #1: Photorhabdus luminescens Makes caterpillars floppy (Mcf) toxin wi...
| Supramolecule | Name: Photorhabdus luminescens Makes caterpillars floppy (Mcf) toxin with the C-terminal deletion type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Photorhabdus luminescens (bacteria) Photorhabdus luminescens (bacteria) |
-Macromolecule #1: Toxin protein
| Macromolecule | Name: Toxin protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Photorhabdus luminescens (bacteria) Photorhabdus luminescens (bacteria) |
| Molecular weight | Theoretical: 325.211562 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MGSSHHHHHH SSGLVPRGSH MASISKDFTN LLNTLIDGQI GAASRQTEWF NMSPDERTDY IKQVDERLQE MQQSTLSVLA AQHFQMQDN PVSVGDQLQT LQKRRQQMTD VPGTPAINAY KQQLDRDILL YRRQQTAMTH FDSTWRKVLV MLGPDDSKPL N ATTLRENA ...String: MGSSHHHHHH SSGLVPRGSH MASISKDFTN LLNTLIDGQI GAASRQTEWF NMSPDERTDY IKQVDERLQE MQQSTLSVLA AQHFQMQDN PVSVGDQLQT LQKRRQQMTD VPGTPAINAY KQQLDRDILL YRRQQTAMTH FDSTWRKVLV MLGPDDSKPL N ATTLRENA VDKQAKLDTE IKRLEQQLTI QVADSTFSQK YVTLFSELQA YKDVNARYNA LLKASATEEA AALGALTKVP QA SDDLPVN ISLLMMEERP GYIRMNVALV NASTDGRFKD FFLENGRLVV LTDGVLNFSF GTAARSLAWQ QQYRLKSEPP SFR SPTYTP IRSVLVKTEF VEKYFANYLV SESTLRGGFK AQLLGNGRKM LLTSVDRKVP NQIGIQVSGQ APNTTITREV PLAS ALSDL INQNADIASF RTIGLEGFRQ SSYHPDRDGL FVNIHELERS VGFAGRQYLL EMPQDNDYLS ATPFGVMSVD GDKVS SSHL SKAQTDTLYQ YNAAFFEKLE QLRSGGMKAS RLFEGSIERT AFVQQLVRLL ERNHITPAGV LAPEYPRDNM RDIKGN NLN KVLWEQAFAA SVWRSRDNDP LLFRLATRLV KNPAVVKVLQ NGYVQSDIAQ ARELLAPLYE QWRTRAVEAE TQRVASA NA AQHPSNPKVH VFDQAEVERS LDDKLLILLL TGPQSLEGTD VQLRPMVEAA LLSNEGRSLR KQILFHALRP VADSFSKA A APVNPHAELG VGKIMINNRL NQPDPYLILN TSSEEQAYRD GSYLIKDDKY RSYNQFRPDF KNDATRYMND LDTPFVGGI SGTTQTVSNV LTELFGGALS VKQYWQFQMA NAAFMIRNGY HSFFETFYVA ARYEPEGADS IGKEMLQMFD KYRVEGSKKA LQGKLYDGV MARVLPIINQ GLSAADEFHP PRFTRIGPRP ALLGQAVKDL ELKAGLTSVG DGFEPRQGSA DIHQFVTDPV L FAKTHTVS AEALVRSGRL PAEGSAQLVK VGSGLYELEY TEQSANDISS SSIPAYFLGY NGPNQANAVP AYVDIPKRTI AG NFLFTGT LSGGSLVVTS LDANTFRVYH DGRVNSSLLY DNVVMAVDYK DYQIAGTAEG LAAAYMQYVN HEWQLVLQRQ EYQ RDGQML RLRLRDDEEP LSIQVADSQV VERNQAQFVA YREQIHQQLK KVATQFEVSI SGVSDGVYTE GEFSPDHPAI AAWA KLCAE VYDRINADTK QLVDKRNKLY ENRRNTIRRD LINQQIKQLN ITLEYYKAQY DTVLREAGFV EQSWLWQQIK AKNGS AAVV RIDDTAIQGG GKQRTDSVGE RYAISEAYQR GARGTGFSDG LRNFREIEIP GVDDKMSALE MKRLFLEGKL TSEQQG ALS GRITETSRAE YIDKVLRQTA VFSEDFHDAG SVFDRLVPQD FYLSLVGDRS GGRAYPLVRA MTVALASGGE AGINSLV QK LFFASADPQA GSSTLLRNSL IKLHSNVEAV QASTELGQFG LSEVVSRLAA TTGTSMFALN TQNHSMMVGS TVTTEGRR Y YFYDPNVGIF AFDNTKSLSR AMEQHLVGRR LAVHYGSFGS KSAPAFNLIE IDTGKMAEVP VGNGLNVADL TRFEELSSV IGQRRQVEQV MSAQERITED LQLSTALQAF DAEQWGARFE AASTRLAQEH QLDSRWLPII ATTEEQGEGR YRVQFINRDQ PEQTRWLDT DDSTFVEFRR FVDEHMSVLN EHFTLESGRM RPRGGVGEAA PVDGLNAGFA VQALIQWFSD KNRHDAANGM A SPDLATAL KVHSYLNFVQ MVHGGVQDVI KVTALVRTAL RGEVVAAQTS FKEFALSLGH TVNEGVGVLF GGAMIGLDAY EL AHAENDV QKAVFGTQLA FDSASFVTGA AGIGAGLVGA STAGAVLGGA GVILGGLAVG FTALAQAFGA VAEDAKAVGR YFD TVDKAY KGNGYRYDNE KQVLVPLAGA VIKTLDLSKN QIDFDSQYIY RTHSGSTGSG KINYFFWVGD FPRMVHDRGQ AIEV RSGIG YKDVSRPLEH GDSNVVILPG TPKSYISYEY MLLPGATTRH DAGFDVIRRL EEDKRFDYDF YIFPGEETIR RIHHE YVDT PIEVVLDQRN RQLVAPELPK ELHGFLCYEI KGAGGEYLIG LNEGAKVNLT SDVASTWIID SSQLASDSIS VSKDQL LVG EKGKEVVVKL YLAQNSQVLV VNGKGEVRKV DFTSLTAQVI SEDASKWQVP GQQIEQHLSD LAKAHQLHGQ YVVVENY RH QGRDVGRAFY DVTKDRMLFT DTTNEQAKRA QLGAVMGDYA YFYDADNAVA WRVDIATGQV DAQFEPWFNQ NAGHISRF W QEGDVVYLAR RYRLKEREAE LGYRIIGDRM ELVSAVGDDA LLQLSARIGR HGDELEAILQ GYRSNSTQRG TLMYTLGAR LIQPTSAALV TVFGVDAAGV PHRYWIRTSD GTLIKPNLAP PADQTLHFEA HEQTRSAWQI PADLVLAGSM PLLGGKEVFF FYSKEQKTL FRQEGPGQEV LDANQPSALR VTTPALTNVI NLNGHLVVVT EDGRVARLDA LGQLSYAAVN EHWLKGRIHW W QDLTSVTD GRATLAVFGV KDTDGKSLLP VWYHNGQVVV ASAALQDKHP QFLGFEVDGS SARLFEPASG KLYRQPAMTA DA LAAAFGT DEVLEASAQL PAANELEPEL HLKAAEQVDA GLRLTTVKGE ILLRTHDGKL QLVAVDKDWQ QDNLVRLSQA LAE VAGQWR VKGVLTLQGD DTQGWFDVGS GQVFSIGGIP ATDNLRFIGI AVGKKGAYVY NPTDQMLYQV KESGAQKLNH YADV ERIGS SLLLQDGGKG DLSPMLIAGV DSVVLHGGAG SDTYRLSQTM WSYYRTVVID NDDPNQVLDR LIILAVDAEK IFVSR HEDD LMLTDSVNGT VLVIRKVFGS QAVTHRHLQI DLEGSSSVIS VDHLVKGFTR UniProtKB:  Toxin protein Toxin protein |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: C-flat-2/1 / Material: COPPER / Mesh: 400 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm Bright-field microscopy / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 9032 / Average electron dose: 60.85 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-8p51: |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)