[English] 日本語
 Yorodumi
Yorodumi- EMDB-15084: cryo-EM structure of thioredoxin glutathione reductase in complex... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryo-EM structure of thioredoxin glutathione reductase in complex with a non-competitive inhibitor | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  inhibitor / inhibitor /  complex / flavoreductase / complex / flavoreductase /  FLAVOPROTEIN FLAVOPROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information thioredoxin-disulfide reductase / thioredoxin-disulfide reductase (NADPH) activity / cell redox homeostasis / thioredoxin-disulfide reductase / thioredoxin-disulfide reductase (NADPH) activity / cell redox homeostasis /  flavin adenine dinucleotide binding / flavin adenine dinucleotide binding /  metal ion binding metal ion bindingSimilarity search - Function | |||||||||
| Biological species |   Schistosoma mansoni (invertebrata) Schistosoma mansoni (invertebrata) | |||||||||
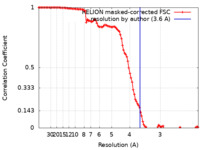
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.6 Å cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Ardini M / Angelucci F / Fata F / Gabriele F / Effantin G / Ling W / Williams DL / Petukhova VZ / Petukhov PA | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Non-covalent inhibitors of thioredoxin glutathione reductase with schistosomicidal activity in vivo. Authors: Valentina Z Petukhova / Sammy Y Aboagye / Matteo Ardini / Rachel P Lullo / Francesca Fata / Margaret E Byrne / Federica Gabriele / Lucy M Martin / Luke N M Harding / Vamshikrishna Gone / ...Authors: Valentina Z Petukhova / Sammy Y Aboagye / Matteo Ardini / Rachel P Lullo / Francesca Fata / Margaret E Byrne / Federica Gabriele / Lucy M Martin / Luke N M Harding / Vamshikrishna Gone / Bikash Dangi / Daniel D Lantvit / Dejan Nikolic / Rodolfo Ippoliti / Grégory Effantin / Wai Li Ling / Jeremy J Johnson / Gregory R J Thatcher / Francesco Angelucci / David L Williams / Pavel A Petukhov /    Abstract: Only praziquantel is available for treating schistosomiasis, a disease affecting more than 200 million people. Praziquantel-resistant worms have been selected for in the lab and low cure rates from ...Only praziquantel is available for treating schistosomiasis, a disease affecting more than 200 million people. Praziquantel-resistant worms have been selected for in the lab and low cure rates from mass drug administration programs suggest that resistance is evolving in the field. Thioredoxin glutathione reductase (TGR) is essential for schistosome survival and a validated drug target. TGR inhibitors identified to date are irreversible and/or covalent inhibitors with unacceptable off-target effects. In this work, we identify noncovalent TGR inhibitors with efficacy against schistosome infections in mice, meeting the criteria for lead progression indicated by WHO. Comparisons with previous in vivo studies with praziquantel suggests that these inhibitors outperform the drug of choice for schistosomiasis against juvenile worms. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15084.map.gz emd_15084.map.gz | 94.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15084-v30.xml emd-15084-v30.xml emd-15084.xml emd-15084.xml | 22.6 KB 22.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15084_fsc.xml emd_15084_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_15084.png emd_15084.png | 43.3 KB | ||
| Others |  emd_15084_additional_1.map.gz emd_15084_additional_1.map.gz emd_15084_half_map_1.map.gz emd_15084_half_map_1.map.gz emd_15084_half_map_2.map.gz emd_15084_half_map_2.map.gz | 96 MB 95.8 MB 95.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15084 http://ftp.pdbj.org/pub/emdb/structures/EMD-15084 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15084 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15084 | HTTPS FTP |
-Related structure data
| Related structure data |  8a1rMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15084.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15084.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.145 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: This is the final sharpened map made by...
| File | emd_15084_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is the final sharpened map made by Phenix (version 1.19.2-4158) local anisotropic sharpening from the refined fullmap_handcoot_map.ccp4 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_15084_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_15084_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : TGR in complex with an inhibitor
| Entire | Name: TGR in complex with an inhibitor |
|---|---|
| Components |
|
-Supramolecule #1: TGR in complex with an inhibitor
| Supramolecule | Name: TGR in complex with an inhibitor / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Recombinant TGR bound non-covalently to a synthetic chimeric compound |
|---|---|
| Source (natural) | Organism:   Schistosoma mansoni (invertebrata) Schistosoma mansoni (invertebrata) |
-Supramolecule #2: TGR complex with inhibitor
| Supramolecule | Name: TGR complex with inhibitor / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Schistosoma mansoni (invertebrata) Schistosoma mansoni (invertebrata) |
-Macromolecule #1: Thioredoxin glutathione reductase
| Macromolecule | Name: Thioredoxin glutathione reductase / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: ec: 1.6.4.5 |
|---|---|
| Source (natural) | Organism:   Schistosoma mansoni (invertebrata) Schistosoma mansoni (invertebrata) |
| Molecular weight | Theoretical: 65.061145 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: MPPADGTSQW LRKTVDSAAV ILFSKTTCPY CKKVKDVLAE AKIKHATIEL DQLSNGSAIQ KCLASFSKIE TVPQMFVRGK FIGDSQTVL KYYSNDELAG IVNESKYDYD LIVIGGGSGG LAAGKEAAKY GAKTAVLDYV EPTPIGTTWG LGGTCVNVGC I PKKLMHQA ...String: MPPADGTSQW LRKTVDSAAV ILFSKTTCPY CKKVKDVLAE AKIKHATIEL DQLSNGSAIQ KCLASFSKIE TVPQMFVRGK FIGDSQTVL KYYSNDELAG IVNESKYDYD LIVIGGGSGG LAAGKEAAKY GAKTAVLDYV EPTPIGTTWG LGGTCVNVGC I PKKLMHQA GLLSHALEDA EHFGWSLDRS KISHNWSTMV EGVQSHIGSL NWGYKVALRD NQVTYLNAKG RLISPHEVQI TD KNQKVST ITGNKIILAT GERPKYPEIP GAVEYGITSD DLFSLPYFPG KTLVIGASYV ALECAGFLAS LGGDVTVMVR SIL LRGFDQ QMAEKVGDYM ENHGVKFAKL CVPDEIKQLK VVDTENNKPG LLLVKGHYTD GKKFEEEFET VIFAVGREPQ LSKV LCETV GVKLDKNGRV VCTDDEQTTV SNVYAIGDIN AGKPQLTPVA IQAGRYLARR LFAGATELTD YSNVATTVFT PLEYG ACGL SEEDAIEKYG DKDIEVYHSN FKPLEWTVAH REDNVCYMKL VCRKSDNMRV LGLHVLGPNA GEITQGYAVA IKMGAT KAD FDRTIGIHPT CSETFTTLHV TKKSGVSPIV SGCCG |
-Macromolecule #2: FLAVIN-ADENINE DINUCLEOTIDE
| Macromolecule | Name: FLAVIN-ADENINE DINUCLEOTIDE / type: ligand / ID: 2 / Number of copies: 2 / Formula: FAD |
|---|---|
| Molecular weight | Theoretical: 785.55 Da |
| Chemical component information |  ChemComp-FAD: |
-Macromolecule #3: (2~{R},3~{R},4~{S},5~{R})-2-[3-[[[(1~{R},2~{R},3~{R},5~{S})-2,6,6...
| Macromolecule | Name: (2~{R},3~{R},4~{S},5~{R})-2-[3-[[[(1~{R},2~{R},3~{R},5~{S})-2,6,6-trimethyl-3-bicyclo[3.1.1]heptanyl]amino]methyl]indol-1-yl]oxane-3,4,5-triol type: ligand / ID: 3 / Number of copies: 2 / Formula: KW2 |
|---|---|
| Molecular weight | Theoretical: 414.538 Da |
| Chemical component information | 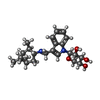 ChemComp-KW2: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: filtered aqueous fresh solution | |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY ARRAY / Support film - Film thickness: 50 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. / Pretreatment - Atmosphere: OTHER | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293.15 K / Instrument: FEI VITROBOT MARK IV / Details: Blotting time= 7s, wait time= 10 s. | |||||||||
| Details | Mono-dispersed TGR molecules in aqueous sample buffer containing DMSO and inhibitor |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 150.0 µm / Illumination mode: SPOT SCAN / Imaging mode: DIFFRACTION / Cs: 2.7 mm / Nominal defocus max: 2.6 µm / Nominal defocus min: 1.8 µm / Nominal magnification: 12000 / Cs: 2.7 mm / Nominal defocus max: 2.6 µm / Nominal defocus min: 1.8 µm / Nominal magnification: 12000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Details | Manual preliminar screening |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Digitization - Frames/image: 1-50 / Number grids imaged: 1 / Number real images: 2600 / Average electron dose: 50.0 e/Å2 |
 Movie
Movie Controller
Controller






 Z
Z Y
Y X
X