+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of P. luminescens TccC3-F-actin complex | |||||||||
 Map data Map data | The map was used for building the model | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationcytoskeletal motor activator activity / detection of maltose stimulus /  tropomyosin binding / tropomyosin binding /  myosin heavy chain binding / myosin heavy chain binding /  maltose binding / maltose transport complex / mesenchyme migration / maltose transport / maltodextrin transmembrane transport / maltose binding / maltose transport complex / mesenchyme migration / maltose transport / maltodextrin transmembrane transport /  troponin I binding ...cytoskeletal motor activator activity / detection of maltose stimulus / troponin I binding ...cytoskeletal motor activator activity / detection of maltose stimulus /  tropomyosin binding / tropomyosin binding /  myosin heavy chain binding / myosin heavy chain binding /  maltose binding / maltose transport complex / mesenchyme migration / maltose transport / maltodextrin transmembrane transport / maltose binding / maltose transport complex / mesenchyme migration / maltose transport / maltodextrin transmembrane transport /  troponin I binding / actin filament bundle / filamentous actin / skeletal muscle thin filament assembly / actin filament bundle assembly / striated muscle thin filament / carbohydrate transmembrane transporter activity / skeletal muscle myofibril / actin monomer binding / ATP-binding cassette (ABC) transporter complex, substrate-binding subunit-containing / carbohydrate transport / skeletal muscle fiber development / troponin I binding / actin filament bundle / filamentous actin / skeletal muscle thin filament assembly / actin filament bundle assembly / striated muscle thin filament / carbohydrate transmembrane transporter activity / skeletal muscle myofibril / actin monomer binding / ATP-binding cassette (ABC) transporter complex, substrate-binding subunit-containing / carbohydrate transport / skeletal muscle fiber development /  stress fiber / stress fiber /  titin binding / actin filament polymerization / ATP-binding cassette (ABC) transporter complex / cell chemotaxis / titin binding / actin filament polymerization / ATP-binding cassette (ABC) transporter complex / cell chemotaxis /  filopodium / filopodium /  actin filament / actin filament /  Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / calcium-dependent protein binding / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / calcium-dependent protein binding /  lamellipodium / lamellipodium /  cell body / outer membrane-bounded periplasmic space / cell body / outer membrane-bounded periplasmic space /  periplasmic space / periplasmic space /  hydrolase activity / protein domain specific binding / DNA damage response / hydrolase activity / protein domain specific binding / DNA damage response /  calcium ion binding / positive regulation of gene expression / magnesium ion binding / calcium ion binding / positive regulation of gene expression / magnesium ion binding /  ATP binding / ATP binding /  membrane / identical protein binding / membrane / identical protein binding /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Photorhabdus luminescens (bacteria) / Photorhabdus luminescens (bacteria) /   rabbit (rabbit) rabbit (rabbit) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.8 Å cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Belyy A / Raunser S | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Mechanism of threonine ADP-ribosylation of F-actin by a Tc toxin. Authors: Alexander Belyy / Florian Lindemann / Daniel Roderer / Johanna Funk / Benjamin Bardiaux / Jonas Protze / Peter Bieling / Hartmut Oschkinat / Stefan Raunser /   Abstract: Tc toxins deliver toxic enzymes into host cells by a unique injection mechanism. One of these enzymes is the actin ADP-ribosyltransferase TccC3, whose activity leads to the clustering of the cellular ...Tc toxins deliver toxic enzymes into host cells by a unique injection mechanism. One of these enzymes is the actin ADP-ribosyltransferase TccC3, whose activity leads to the clustering of the cellular cytoskeleton and ultimately cell death. Here, we show in atomic detail how TccC3 modifies actin. We find that the ADP-ribosyltransferase does not bind to G-actin but interacts with two consecutive actin subunits of F-actin. The binding of TccC3 to F-actin occurs via an induced-fit mechanism that facilitates access of NAD to the nucleotide binding pocket. The following nucleophilic substitution reaction results in the transfer of ADP-ribose to threonine-148 of F-actin. We demonstrate that this site-specific modification of F-actin prevents its interaction with depolymerization factors, such as cofilin, which impairs actin network turnover and leads to steady actin polymerization. Our findings reveal in atomic detail a mechanism of action of a bacterial toxin through specific targeting and modification of F-actin. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14532.map.gz emd_14532.map.gz | 9.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14532-v30.xml emd-14532-v30.xml emd-14532.xml emd-14532.xml | 18.7 KB 18.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_14532.png emd_14532.png | 136.2 KB | ||
| Masks |  emd_14532_msk_1.map emd_14532_msk_1.map | 125 MB |  Mask map Mask map | |
| Others |  emd_14532_half_map_1.map.gz emd_14532_half_map_1.map.gz emd_14532_half_map_2.map.gz emd_14532_half_map_2.map.gz | 60.1 MB 60.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14532 http://ftp.pdbj.org/pub/emdb/structures/EMD-14532 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14532 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14532 | HTTPS FTP |
-Related structure data
| Related structure data |  7z7hMC  7z7iC  7zbqC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14532.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14532.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The map was used for building the model | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.9 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14532_msk_1.map emd_14532_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Second half map
| File | emd_14532_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Second half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: First half map
| File | emd_14532_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | First half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Structure of P. luminescence TccC3-F-actin complex
| Entire | Name: Structure of P. luminescence TccC3-F-actin complex |
|---|---|
| Components |
|
-Supramolecule #1: Structure of P. luminescence TccC3-F-actin complex
| Supramolecule | Name: Structure of P. luminescence TccC3-F-actin complex / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Photorhabdus luminescens (bacteria) Photorhabdus luminescens (bacteria) |
-Macromolecule #1: Actin, alpha skeletal muscle
| Macromolecule | Name: Actin, alpha skeletal muscle / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   rabbit (rabbit) rabbit (rabbit) |
| Molecular weight | Theoretical: 42.109973 KDa |
| Sequence | String: MCDEDETTAL VCDNGSGLVK AGFAGDDAPR AVFPSIVGRP RHQGVMVGMG QKDSYVGDEA QSKRGILTLK YPIE(HIC)G IIT NWDDMEKIWH HTFYNELRVA PEEHPTLLTE APLNPKANRE KMTQIMFETF NVPAMYVAIQ AVLSLYASGR TTGIVLD SG DGVTHNVPIY ...String: MCDEDETTAL VCDNGSGLVK AGFAGDDAPR AVFPSIVGRP RHQGVMVGMG QKDSYVGDEA QSKRGILTLK YPIE(HIC)G IIT NWDDMEKIWH HTFYNELRVA PEEHPTLLTE APLNPKANRE KMTQIMFETF NVPAMYVAIQ AVLSLYASGR TTGIVLD SG DGVTHNVPIY EGYALPHAIM RLDLAGRDLT DYLMKILTER GYSFVTTAER EIVRDIKEKL CYVALDFENE MATAASSS S LEKSYELPDG QVITIGNERF RCPETLFQPS FIGMESAGIH ETTYNSIMKC DIDIRKDLYA NNVMSGGTTM YPGIADRMQ KEITALAPST MKIKIIAPPE RKYSVWIGGS ILASLSTFQQ MWITKQEYDE AGPSIVHRKC F |
-Macromolecule #2: Maltose/maltodextrin-binding periplasmic protein,TccC3
| Macromolecule | Name: Maltose/maltodextrin-binding periplasmic protein,TccC3 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Photorhabdus luminescens (bacteria) Photorhabdus luminescens (bacteria) |
| Molecular weight | Theoretical: 63.658949 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21(DE3) (bacteria) Escherichia coli BL21(DE3) (bacteria) |
| Sequence | String: MGSSHHHHHH SSGLVPRGSH MKIEEGKLVI WINGDKGYNG LAEVGKKFEK DTGIKVTVEH PDKLEEKFPQ VAATGDGPDI IFWAHDRFG GYAQSGLLAE ITPDKAFQDK LYPFTWDAVR YNGKLIAYPI AVEALSLIYN KDLLPNPPKT WEEIPALDKE L KAKGKSAL ...String: MGSSHHHHHH SSGLVPRGSH MKIEEGKLVI WINGDKGYNG LAEVGKKFEK DTGIKVTVEH PDKLEEKFPQ VAATGDGPDI IFWAHDRFG GYAQSGLLAE ITPDKAFQDK LYPFTWDAVR YNGKLIAYPI AVEALSLIYN KDLLPNPPKT WEEIPALDKE L KAKGKSAL MFNLQEPYFT WPLIAADGGY AFKYENGKYD IKDVGVDNAG AKAGLTFLVD LIKNKHMNAD TDYSIAEAAF NK GETAMTI NGPWAWSNID TSKVNYGVTV LPTFKGQPSK PFVGVLSAGI NAASPNKELA KEFLENYLLT DEGLEAVNKD KPL GAVALK SYEEELVKDP RIAATMENAQ KGEIMPNIPQ MSAFWYAVRT AVINAASGRQ TVDEALKDAQ TNSGSSGSSG STNL QKKSF TLYRADNRSF EEMQSKFPEG FKAWTPLDTK MARQFASIFI GQKDTSNLPK ETVKNISTWG AKPKLKDLSN YIKYT KDKS TVWVSTAINT EAGGQSSGAP LHKIDMDLYE FAIDGQKLNP LPEGRTKNMV PSLLLDTPQI ETSSIIALNH GPVNDA SIS FLTTIPLKNV KPHKR |
-Macromolecule #3: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 5 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 5 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #5: ADENOSINE-5-DIPHOSPHORIBOSE
| Macromolecule | Name: ADENOSINE-5-DIPHOSPHORIBOSE / type: ligand / ID: 5 / Number of copies: 1 / Formula: APR |
|---|---|
| Molecular weight | Theoretical: 559.316 Da |
| Chemical component information | 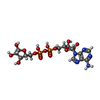 ChemComp-APR: |
-Macromolecule #6: NICOTINAMIDE
| Macromolecule | Name: NICOTINAMIDE / type: ligand / ID: 6 / Number of copies: 1 / Formula: NCA |
|---|---|
| Molecular weight | Theoretical: 122.125 Da |
| Chemical component information | 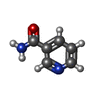 ChemComp-NCA: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 0.4 µm Bright-field microscopy / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 0.4 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 12284 / Average electron dose: 84.9 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Particle selection | Number selected: 2224805 |
|---|---|
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: SPHIRE |
| Final 3D classification | Software - Name: RELION (ver. 3) |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: SPHIRE |
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: FOURIER SPACE / Resolution.type: BY AUTHOR / Resolution: 3.8 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: SPHIRE / Number images used: 437658 |
 Movie
Movie Controller
Controller











 Z
Z Y
Y X
X