[English] 日本語
 Yorodumi
Yorodumi- EMDB-12257: Plasmodium falciparum kinesin-5 motor domain without nucleotide, ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12257 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Plasmodium falciparum kinesin-5 motor domain without nucleotide, complexed with 14 protofilament microtubule. | |||||||||||||||
 Map data Map data | Asymmetric unit of Plasmodium falciparum kinesin-5 motor domain without nucleotide, in complex with porcine 14 protofilament microtubules. | |||||||||||||||
 Sample Sample |
| |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationKinesins / Microtubule-dependent trafficking of connexons from Golgi to the plasma membrane / Hedgehog 'off' state /  Cilium Assembly / Cilium Assembly /  Intraflagellar transport / COPI-dependent Golgi-to-ER retrograde traffic / Carboxyterminal post-translational modifications of tubulin / RHOH GTPase cycle / Sealing of the nuclear envelope (NE) by ESCRT-III / Kinesins ...Kinesins / Microtubule-dependent trafficking of connexons from Golgi to the plasma membrane / Hedgehog 'off' state / Intraflagellar transport / COPI-dependent Golgi-to-ER retrograde traffic / Carboxyterminal post-translational modifications of tubulin / RHOH GTPase cycle / Sealing of the nuclear envelope (NE) by ESCRT-III / Kinesins ...Kinesins / Microtubule-dependent trafficking of connexons from Golgi to the plasma membrane / Hedgehog 'off' state /  Cilium Assembly / Cilium Assembly /  Intraflagellar transport / COPI-dependent Golgi-to-ER retrograde traffic / Carboxyterminal post-translational modifications of tubulin / RHOH GTPase cycle / Sealing of the nuclear envelope (NE) by ESCRT-III / Kinesins / PKR-mediated signaling / Resolution of Sister Chromatid Cohesion / Mitotic Prometaphase / EML4 and NUDC in mitotic spindle formation / Separation of Sister Chromatids / The role of GTSE1 in G2/M progression after G2 checkpoint / Aggrephagy / plus-end-directed microtubule motor activity / Recruitment of NuMA to mitotic centrosomes / RHO GTPases activate IQGAPs / RHO GTPases Activate Formins / HSP90 chaperone cycle for steroid hormone receptors (SHR) in the presence of ligand / COPI-independent Golgi-to-ER retrograde traffic / MHC class II antigen presentation / COPI-mediated anterograde transport / Intraflagellar transport / COPI-dependent Golgi-to-ER retrograde traffic / Carboxyterminal post-translational modifications of tubulin / RHOH GTPase cycle / Sealing of the nuclear envelope (NE) by ESCRT-III / Kinesins / PKR-mediated signaling / Resolution of Sister Chromatid Cohesion / Mitotic Prometaphase / EML4 and NUDC in mitotic spindle formation / Separation of Sister Chromatids / The role of GTSE1 in G2/M progression after G2 checkpoint / Aggrephagy / plus-end-directed microtubule motor activity / Recruitment of NuMA to mitotic centrosomes / RHO GTPases activate IQGAPs / RHO GTPases Activate Formins / HSP90 chaperone cycle for steroid hormone receptors (SHR) in the presence of ligand / COPI-independent Golgi-to-ER retrograde traffic / MHC class II antigen presentation / COPI-mediated anterograde transport /  kinesin complex / kinesin complex /  microtubule motor activity / microtubule-based movement / microtubule motor activity / microtubule-based movement /  Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / structural constituent of cytoskeleton / microtubule cytoskeleton organization / microtubule cytoskeleton / mitotic cell cycle / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / structural constituent of cytoskeleton / microtubule cytoskeleton organization / microtubule cytoskeleton / mitotic cell cycle /  microtubule binding / microtubule binding /  microtubule / microtubule /  GTPase activity / GTP binding / GTPase activity / GTP binding /  ATP binding / ATP binding /  metal ion binding / metal ion binding /  cytoplasm cytoplasmSimilarity search - Function | |||||||||||||||
| Biological species |   Sus scrofa (pig) / Sus scrofa (pig) /   Plasmodium falciparum (isolate 3D7) (eukaryote) / Plasmodium falciparum (isolate 3D7) (eukaryote) /   Pig (pig) Pig (pig) | |||||||||||||||
| Method | helical reconstruction /  cryo EM / Resolution: 4.4 Å cryo EM / Resolution: 4.4 Å | |||||||||||||||
 Authors Authors | Cook AD / Roberts A / Atherton J / Tewari R / Topf M / Moores CA | |||||||||||||||
| Funding support |  United Kingdom, 4 items United Kingdom, 4 items
| |||||||||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2021 Journal: J Biol Chem / Year: 2021Title: Cryo-EM structure of a microtubule-bound parasite kinesin motor and implications for its mechanism and inhibition. Authors: Alexander D Cook / Anthony J Roberts / Joseph Atherton / Rita Tewari / Maya Topf / Carolyn A Moores /  Abstract: Plasmodium parasites cause malaria and are responsible annually for hundreds of thousands of deaths. Kinesins are a superfamily of microtubule-dependent ATPases that play important roles in the ...Plasmodium parasites cause malaria and are responsible annually for hundreds of thousands of deaths. Kinesins are a superfamily of microtubule-dependent ATPases that play important roles in the parasite replicative machinery, which is a potential target for antiparasite drugs. Kinesin-5, a molecular motor that cross-links microtubules, is an established antimitotic target in other disease contexts, but its mechanism in Plasmodium falciparum is unclear. Here, we characterized P. falciparum kinesin-5 (PfK5) using cryo-EM to determine the motor's nucleotide-dependent microtubule-bound structure and introduced 3D classification of individual motors into our microtubule image processing pipeline to maximize our structural insights. Despite sequence divergence in PfK5, the motor exhibits classical kinesin mechanochemistry, including ATP-induced subdomain rearrangement and cover neck bundle formation, consistent with its plus-ended directed motility. We also observed that an insertion in loop5 of the PfK5 motor domain creates a different environment in the well-characterized human kinesin-5 drug-binding site. Our data reveal the possibility for selective inhibition of PfK5 and can be used to inform future exploration of Plasmodium kinesins as antiparasite targets. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12257.map.gz emd_12257.map.gz | 370.7 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12257-v30.xml emd-12257-v30.xml emd-12257.xml emd-12257.xml | 23.6 KB 23.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12257_fsc.xml emd_12257_fsc.xml | 15.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_12257.png emd_12257.png | 106.9 KB | ||
| Others |  emd_12257_additional_1.map.gz emd_12257_additional_1.map.gz emd_12257_additional_2.map.gz emd_12257_additional_2.map.gz | 170.9 MB 167.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12257 http://ftp.pdbj.org/pub/emdb/structures/EMD-12257 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12257 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12257 | HTTPS FTP |
-Related structure data
| Related structure data |  7nb8MC  7nbaC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12257.map.gz / Format: CCP4 / Size: 1.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12257.map.gz / Format: CCP4 / Size: 1.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Asymmetric unit of Plasmodium falciparum kinesin-5 motor domain without nucleotide, in complex with porcine 14 protofilament microtubules. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.39 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Asymmetric (C1) reconstruction of Plasmodium falciparum kinesin-5 motor...
| File | emd_12257_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Asymmetric (C1) reconstruction of Plasmodium falciparum kinesin-5 motor domain without nucleotide, in complex with porcine 14 protofilament microtubules. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Symmetry expanded reconstruction of Plasmodium falciparum kinesin-5 motor...
| File | emd_12257_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Symmetry expanded reconstruction of Plasmodium falciparum kinesin-5 motor without nucleotide, in complex with porcine 14 protofilament microtubules. After motor domain 3D classification. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : 14 protofilament microtubule of alpha/beta-tubulin dimers, with k...
| Entire | Name: 14 protofilament microtubule of alpha/beta-tubulin dimers, with kinesin-5 motor domain bound 1:1 to tubulin dimers. |
|---|---|
| Components |
|
-Supramolecule #1: 14 protofilament microtubule of alpha/beta-tubulin dimers, with k...
| Supramolecule | Name: 14 protofilament microtubule of alpha/beta-tubulin dimers, with kinesin-5 motor domain bound 1:1 to tubulin dimers. type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|
-Supramolecule #2: alpha/beta-tubulin
| Supramolecule | Name: alpha/beta-tubulin / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Sus scrofa (pig) Sus scrofa (pig) |
-Supramolecule #3: Kinesin-5
| Supramolecule | Name: Kinesin-5 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism:   Plasmodium falciparum (isolate 3D7) (eukaryote) Plasmodium falciparum (isolate 3D7) (eukaryote) |
| Recombinant expression | Organism:   Escherichia coli BL21 (bacteria) Escherichia coli BL21 (bacteria) |
-Macromolecule #1: Tubulin alpha-1B chain
| Macromolecule | Name: Tubulin alpha-1B chain / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Pig (pig) / Organ: Brain Pig (pig) / Organ: Brain |
| Molecular weight | Theoretical: 50.204445 KDa |
| Sequence | String: MRECISIHVG QAGVQIGNAC WELYCLEHGI QPDGQMPSDK TIGGGDDSFN TFFSETGAGK HVPRAVFVDL EPTVIDEVRT GTYRQLFHP EQLITGKEDA ANNYARGHYT IGKEIIDLVL DRIRKLADQC TGLQGFLVFH SFGGGTGSGF TSLLMERLSV D YGKKSKLE ...String: MRECISIHVG QAGVQIGNAC WELYCLEHGI QPDGQMPSDK TIGGGDDSFN TFFSETGAGK HVPRAVFVDL EPTVIDEVRT GTYRQLFHP EQLITGKEDA ANNYARGHYT IGKEIIDLVL DRIRKLADQC TGLQGFLVFH SFGGGTGSGF TSLLMERLSV D YGKKSKLE FSIYPAPQVS TAVVEPYNSI LTTHTTLEHS DCAFMVDNEA IYDICRRNLD IERPTYTNLN RLISQIVSSI TA SLRFDGA LNVDLTEFQT NLVPYPRIHF PLATYAPVIS AEKAYHEQLS VAEITNACFE PANQMVKCDP RHGKYMACCL LYR GDVVPK DVNAAIATIK TKRSIQFVDW CPTGFKVGIN YQPPTVVPGG DLAKVQRAVC MLSNTTAIAE AWARLDHKFD LMYA KRAFV HWYVGEGMEE GEFSEAREDM AALEKDYEEV GVDSVEGEGE EEGEEY |
-Macromolecule #2: Tubulin beta chain
| Macromolecule | Name: Tubulin beta chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Pig (pig) / Organ: Brain Pig (pig) / Organ: Brain |
| Molecular weight | Theoretical: 49.90777 KDa |
| Sequence | String: MREIVHIQAG QCGNQIGAKF WEVISDEHGI DPTGSYHGDS DLQLERINVY YNEAAGNKYV PRAILVDLEP GTMDSVRSGP FGQIFRPDN FVFGQSGAGN NWAKGHYTEG AELVDSVLDV VRKESESCDC LQGFQLTHSL GGGTGSGMGT LLISKIREEY P DRIMNTFS ...String: MREIVHIQAG QCGNQIGAKF WEVISDEHGI DPTGSYHGDS DLQLERINVY YNEAAGNKYV PRAILVDLEP GTMDSVRSGP FGQIFRPDN FVFGQSGAGN NWAKGHYTEG AELVDSVLDV VRKESESCDC LQGFQLTHSL GGGTGSGMGT LLISKIREEY P DRIMNTFS VVPSPKVSDT VVEPYNATLS VHQLVENTDE TYCIDNEALY DICFRTLKLT TPTYGDLNHL VSATMSGVTT CL RFPGQLN ADLRKLAVNM VPFPRLHFFM PGFAPLTSRG SQQYRALTVP ELTQQMFDAK NMMAACDPRH GRYLTVAAVF RGR MSMKEV DEQMLNVQNK NSSYFVEWIP NNVKTAVCDI PPRGLKMSAT FIGNSTAIQE LFKRISEQFT AMFRRKAFLH WYTG EGMDE MEFTEAESNM NDLVSEYQQY QDATADEQGE FEEEGEEDEA |
-Macromolecule #3: Kinesin-5
| Macromolecule | Name: Kinesin-5 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Plasmodium falciparum (isolate 3D7) (eukaryote) / Strain: isolate 3D7 Plasmodium falciparum (isolate 3D7) (eukaryote) / Strain: isolate 3D7 |
| Molecular weight | Theoretical: 46.910113 KDa |
| Recombinant expression | Organism:   Escherichia coli BL21 (bacteria) Escherichia coli BL21 (bacteria) |
| Sequence | String: GIDPFTMLRN SYNNDKSSCV NIKVIVRCRP LNEKEKNDIN NEEVVRINNN EVILTINRNN EIYEKKYSFD YACDKDVDQK TLFNNYIYQ IVDEVLQGFN CTLFCYGQTG TGKTYTMEGK ILEHLKQYDN NKKVDLNESI NSDISYCYEL CENEDTGLIF R VTKRIFDI ...String: GIDPFTMLRN SYNNDKSSCV NIKVIVRCRP LNEKEKNDIN NEEVVRINNN EVILTINRNN EIYEKKYSFD YACDKDVDQK TLFNNYIYQ IVDEVLQGFN CTLFCYGQTG TGKTYTMEGK ILEHLKQYDN NKKVDLNESI NSDISYCYEL CENEDTGLIF R VTKRIFDI LNKRKEEKIR HFDKNMYDFN IKISYLEIYN EELCDLLSST NENMKLRIYE DSNNKSKGLN VDKLEEKSIN SF EEIYYII CSAIKKRRTA ETAYNKKSSR SHSIFTITLI IKDINNVGES ITKIGKLNLV DLAGSENALK SSYGSLKIRQ QES CNINQS LLTLGRVINS LIENSSYIPY RDSKLTRLLQ DSLGGKTKTF IVATISPSSL CIDETLSTLD YVFRAKNIKN RPEI NIKTT |
-Macromolecule #4: GUANOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 1 / Formula: GTP |
|---|---|
| Molecular weight | Theoretical: 523.18 Da |
| Chemical component information | 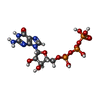 ChemComp-GTP: |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #6: PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER
| Macromolecule | Name: PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER / type: ligand / ID: 6 / Number of copies: 1 / Formula: G2P |
|---|---|
| Molecular weight | Theoretical: 521.208 Da |
| Chemical component information |  ChemComp-G2P: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 3.7 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.0003 kPa | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 296 K / Instrument: FEI VITROBOT MARK IV Details: 3.5 uM microtubules were incubated to the grid and incubated for 30 seconds. The grid was blotted, then 40 uM kinesin motor domain added and incubated for 30 seconds. The grid was again ...Details: 3.5 uM microtubules were incubated to the grid and incubated for 30 seconds. The grid was blotted, then 40 uM kinesin motor domain added and incubated for 30 seconds. The grid was again blotted and 40 uM kinesin motor domain added for 30 seconds. Then the grid was blotted and plunge frozen.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.0 mm Bright-field microscopy / Cs: 2.0 mm |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 335 / Average exposure time: 18.0 sec. / Average electron dose: 58.0 e/Å2 |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




















 Z
Z Y
Y X
X




















