+Search query
-Structure paper
| Title | Orthosteric and allosteric modulation of human HCAR2 signaling complex. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 14, Issue 1, Page 7620, Year 2023 |
| Publish date | Nov 22, 2023 |
 Authors Authors | Chunyou Mao / Mengru Gao / Shao-Kun Zang / Yanqing Zhu / Dan-Dan Shen / Li-Nan Chen / Liu Yang / Zhiwei Wang / Huibing Zhang / Wei-Wei Wang / Qingya Shen / Yanhui Lu / Xin Ma / Yan Zhang /  |
| PubMed Abstract | Hydroxycarboxylic acids are crucial metabolic intermediates involved in various physiological and pathological processes, some of which are recognized by specific hydroxycarboxylic acid receptors ...Hydroxycarboxylic acids are crucial metabolic intermediates involved in various physiological and pathological processes, some of which are recognized by specific hydroxycarboxylic acid receptors (HCARs). HCAR2 is one such receptor, activated by endogenous β-hydroxybutyrate (3-HB) and butyrate, and is the target for Niacin. Interest in HCAR2 has been driven by its potential as a therapeutic target in cardiovascular and neuroinflammatory diseases. However, the limited understanding of how ligands bind to this receptor has hindered the development of alternative drugs able to avoid the common flushing side-effects associated with Niacin therapy. Here, we present three high-resolution structures of HCAR2-Gi1 complexes bound to four different ligands, one potent synthetic agonist (MK-6892) bound alone, and the two structures bound to the allosteric agonist compound 9n in conjunction with either the endogenous ligand 3-HB or niacin. These structures coupled with our functional and computational analyses further our understanding of ligand recognition, allosteric modulation, and activation of HCAR2 and pave the way for the development of high-efficiency drugs with reduced side-effects. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:37993467 / PubMed:37993467 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.55 - 2.76 Å |
| Structure data | EMDB-36010, PDB-8j6p: EMDB-36011, PDB-8j6q: EMDB-36012, PDB-8j6r: |
| Chemicals |  ChemComp-IX8:  ChemComp-CLR: 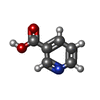 ChemComp-NIO:  ChemComp-NAG:  ChemComp-HOH:  ChemComp-3HR: 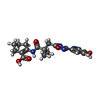 ChemComp-FI7: |
| Source |
|
 Keywords Keywords |  MEMBRANE PROTEIN / hydroxycarboxylic acid receptor / MEMBRANE PROTEIN / hydroxycarboxylic acid receptor /  Niacin / compound 9n / Class A GPCR / 3-HB / MK-6892 Niacin / compound 9n / Class A GPCR / 3-HB / MK-6892 |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers










