+Search query
-Structure paper
| Title | Cryo-EM structures of a LptDE transporter in complex with Pro-macrobodies offer insight into lipopolysaccharide translocation. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 13, Issue 1, Page 1826, Year 2022 |
| Publish date | Apr 5, 2022 |
 Authors Authors | Mathieu Botte / Dongchun Ni / Stephan Schenck / Iwan Zimmermann / Mohamed Chami / Nicolas Bocquet / Pascal Egloff / Denis Bucher / Matilde Trabuco / Robert K Y Cheng / Janine D Brunner / Markus A Seeger / Henning Stahlberg / Michael Hennig /   |
| PubMed Abstract | Lipopolysaccharides are major constituents of the extracellular leaflet in the bacterial outer membrane and form an effective physical barrier for environmental threats and for antibiotics in Gram- ...Lipopolysaccharides are major constituents of the extracellular leaflet in the bacterial outer membrane and form an effective physical barrier for environmental threats and for antibiotics in Gram-negative bacteria. The last step of LPS insertion via the Lpt pathway is mediated by the LptD/E protein complex. Detailed insights into the architecture of LptDE transporter complexes have been derived from X-ray crystallography. However, no structure of a laterally open LptD transporter, a transient state that occurs during LPS release, is available to date. Here, we report a cryo-EM structure of a partially opened LptDE transporter in complex with rigid chaperones derived from nanobodies, at 3.4 Å resolution. In addition, a subset of particles allows to model a structure of a laterally fully opened LptDE complex. Our work offers insights into the mechanism of LPS insertion, provides a structural framework for the development of antibiotics targeting LptD and describes a highly rigid chaperone scaffold to enable structural biology of challenging protein targets. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:35383177 / PubMed:35383177 /  PubMed Central PubMed Central |
| Methods | EM (single particle) / X-ray diffraction |
| Resolution | 2 - 3.4 Å |
| Structure data | EMDB-12990: Cryo-EM structure of N. gonorhoeae LptDE in complex with ProMacrobodies  PDB-7omt: |
| Chemicals | 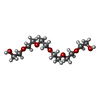 ChemComp-P6G:  ChemComp-MG:  ChemComp-HOH: |
| Source |
|
 Keywords Keywords |  TRANSPORT PROTEIN / Outer membrane protein LPS transporter Bacterial transporter Neisseria gonorrhoeae Drug target ProMacrobodies Structural chaperone Cryo-electron microscopy / TRANSPORT PROTEIN / Outer membrane protein LPS transporter Bacterial transporter Neisseria gonorrhoeae Drug target ProMacrobodies Structural chaperone Cryo-electron microscopy /  IMMUNE SYSTEM / nanobody Cryo-EM Chaperone MBP IMMUNE SYSTEM / nanobody Cryo-EM Chaperone MBP |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers







