+Search query
-Structure paper
| Title | Filament formation drives catalysis by glutaminase enzymes important in cancer progression. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 15, Issue 1, Page 1971, Year 2024 |
| Publish date | Mar 4, 2024 |
 Authors Authors | Shi Feng / Cody Aplin / Thuy-Tien T Nguyen / Shawn K Milano / Richard A Cerione /  |
| PubMed Abstract | The glutaminase enzymes GAC and GLS2 catalyze the hydrolysis of glutamine to glutamate, satisfying the 'glutamine addiction' of cancer cells. They are the targets of anti-cancer drugs; however, their ...The glutaminase enzymes GAC and GLS2 catalyze the hydrolysis of glutamine to glutamate, satisfying the 'glutamine addiction' of cancer cells. They are the targets of anti-cancer drugs; however, their mechanisms of activation and catalytic activity have been unclear. Here we demonstrate that the ability of GAC and GLS2 to form filaments is directly coupled to their catalytic activity and present their cryo-EM structures which provide a view of the conformational states essential for catalysis. Filament formation guides an 'activation loop' to assume a specific conformation that works together with a 'lid' to close over the active site and position glutamine for nucleophilic attack by an essential serine. Our findings highlight how ankyrin repeats on GLS2 regulate enzymatic activity, while allosteric activators stabilize, and clinically relevant inhibitors block, filament formation that enables glutaminases to catalyze glutaminolysis and support cancer progression. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:38438397 / PubMed:38438397 /  PubMed Central PubMed Central |
| Methods | EM (helical sym.) / EM (single particle) |
| Resolution | 3.12 - 3.69 Å |
| Structure data | EMDB-40918, PDB-8szj: EMDB-40920, PDB-8szl: EMDB-40950, PDB-8t0z:  EMDB-43533: Human liver-type glutaminase, bound with inhibitor Compound 968 |
| Chemicals | 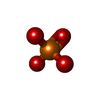 ChemComp-PO4:  ChemComp-GLN: |
| Source |
|
 Keywords Keywords |  HYDROLASE / HYDROLASE /  Cancer / Cancer /  Filament / Filament /  Metabolism / Metabolic Metabolism / Metabolic |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers










