+Search query
-Structure paper
| Title | Molecular insights into the regulation of constitutive activity by RNA editing of 5HT serotonin receptors. |
|---|---|
| Journal, issue, pages | Cell Rep, Vol. 40, Issue 7, Page 111211, Year 2022 |
| Publish date | Aug 16, 2022 |
 Authors Authors | Ryan H Gumpper / Jonathan F Fay / Bryan L Roth /  |
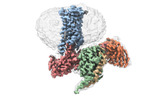
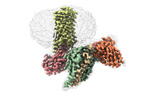
| PubMed Abstract | RNA editing is a process by which post-transcriptional changes of mRNA nucleotides alter protein function through modification of the amino acid content. The 5HT serotonin receptor, which undergoes ...RNA editing is a process by which post-transcriptional changes of mRNA nucleotides alter protein function through modification of the amino acid content. The 5HT serotonin receptor, which undergoes 32 distinct RNA-editing events leading to 24 protein isoforms, is a notable example of this process. These 5HT isoforms display differences in constitutive activity, agonist/inverse agonist potencies, and efficacies. To elucidate the molecular mechanisms responsible for these effects of RNA editing, we present four active-state 5HT-transducer-coupled structures of three representative isoforms (INI, VGV, and VSV) with the selective drug lorcaserin (Belviq) and the classic psychedelic psilocin. We also provide a comprehensive analysis of agonist activation and constitutive activity across all 24 protein isoforms. Collectively, these findings reveal a unique hydrogen-bonding network located on intracellular loop 2 that is subject to RNA editing, which differentially affects GPCR constitutive and agonist signaling activities. |
 External links External links |  Cell Rep / Cell Rep /  PubMed:35977511 / PubMed:35977511 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.84 - 3.6 Å |
| Structure data | EMDB-27633, PDB-8dpf: EMDB-27634, PDB-8dpg: EMDB-27635, PDB-8dph: EMDB-27636, PDB-8dpi: |
| Chemicals |  ChemComp-T4U:  ChemComp-CLR:  ChemComp-91Q: |
| Source |
|
 Keywords Keywords |  MEMBRANE PROTEIN / MEMBRANE PROTEIN /  GPCR GPCR |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers