-Search query
-Search result
Showing all 46 items for (author: fong & k)
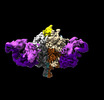
EMDB-41048: 
Lassa GPC Trimer in complex with Fab 8.11G and nanobody D5
Method: single particle / : Gorman J, Kwong PD

PDB-8t5c: 
Lassa GPC Trimer in complex with Fab 8.11G and nanobody D5
Method: single particle / : Gorman J, Kwong PD

EMDB-41302: 
Lassa GPC trimer in complex with Fab GP23
Method: single particle / : Gorman J, Kwong PD

EMDB-34066: 
F1-ATPase of Acinetobacter baumannii
Method: single particle / : Saw WG, Grueber G

EMDB-26859: 
Ligand-free Lassa GPC Trimer with C3 Symmetry
Method: single particle / : Gorman J, Kwong PD
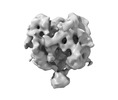
EMDB-26740: 
Ligand-free Lassa GPC Trimer with C1 Symmetry
Method: single particle / : Gorman J, Kwong PD

EMDB-34648: 
CryoEM structure of Helicobacter pylori UreFD/urease complex
Method: single particle / : Nim YS, Fong IYH, Deme J, Tsang KL, Caesar J, Johnson S, Wong KB, Lea SM

EMDB-34659: 
CryoEM Structure of Klebsiella pneumoniae UreD/urease complex
Method: single particle / : Nim YS, Fong IYH, Deme J, Tsang KL, Caesar J, Johnson S, Wong KB, Lea SM

PDB-8hc1: 
CryoEM structure of Helicobacter pylori UreFD/urease complex
Method: single particle / : Nim YS, Fong IYH, Deme J, Tsang KL, Caesar J, Johnson S, Wong KB, Lea SM

PDB-8hcn: 
CryoEM Structure of Klebsiella pneumoniae UreD/urease complex
Method: single particle / : Nim YS, Fong IYH, Deme J, Tsang KL, Caesar J, Johnson S, Wong KB, Lea SM

EMDB-26044: 
H10ssF: ferritin-based nanoparticle displaying H10 hemagglutinin stabilized stem epitopes
Method: single particle / : Gallagher JR, Harris AK

EMDB-22302: 
Cryo-EM Structure of Vaccine-Elicited Rhesus Antibody 789-203-3C12 in Complex with Stabilized SI06 (A/Solomon Islands/3/06) Influenza Hemagglutinin Trimer
Method: single particle / : Cerutti G, Gorman J, Kwong PD, Shapiro L

PDB-6xsk: 
Cryo-EM Structure of Vaccine-Elicited Rhesus Antibody 789-203-3C12 in Complex with Stabilized SI06 (A/Solomon Islands/3/06) Influenza Hemagglutinin Trimer
Method: single particle / : Cerutti G, Gorman J, Kwong PD, Shapiro L

EMDB-21961: 
Cryo-EM structure of the VRC315 clinical trial, vaccine-elicited, human antibody 1D12 in complex with an H7 SH13 HA trimer
Method: single particle / : Gorman J, Kwong PD

PDB-6wxl: 
Cryo-EM structure of the VRC315 clinical trial, vaccine-elicited, human antibody 1D12 in complex with an H7 SH13 HA trimer
Method: single particle / : Gorman J, Kwong PD

EMDB-22804: 
Cryo-EM structure of SRR2899884.46167H+MEDI8852L fab in complex with Victoria HA
Method: single particle / : Gorman J, Kwong PD

PDB-7kc1: 
Cryo-EM structure of SRR2899884.46167H+MEDI8852L fab in complex with Victoria HA
Method: single particle / : Gorman J, Kwong PD

EMDB-10143: 
33mer structure of the Salmonella flagella MS-ring protein FliF
Method: single particle / : Johnson S, Fong YH

EMDB-10145: 
Structure of the RBM3/collar region of the Salmonella flagella MS-ring protein FliF with 33-fold symmetry applied
Method: single particle / : Johnson S, Fong YH

EMDB-10146: 
Structure of the RBM2inner region of the Salmonella flagella MS-ring protein FliF with 21-fold symmetry applied.
Method: single particle / : Johnson S, Fong YH

EMDB-10147: 
34mer structure of the Salmonella flagella MS-ring protein FliF
Method: single particle / : Johnson S, Fong YH

EMDB-10148: 
Structure of the RBM3/collar region of the Salmonella flagella MS-ring protein FliF with 34-fold symmetry applied
Method: single particle / : Johnson S, Fong YH

EMDB-10149: 
Structure of the RBM2 inner ring of Salmonella flagella MS-ring protein FliF with 22-fold symmetry applied
Method: single particle / : Johnson S, Fong YH

EMDB-10560: 
Structure of the RBM3/collar region of the Salmonella flagella MS-ring protein FliF with 32-fold symmetry applied
Method: single particle / : Johnson S, Fong YH

EMDB-10561: 
Structure of the RBM3/collar region of the Salmonella flagella MS-ring protein FliF with 35-fold symmetry applied
Method: single particle / : Johnson S, Fong YH, Deme JC, Furlong EJ, Kuhlen L, Lea SM

PDB-6scn: 
33mer structure of the Salmonella flagella MS-ring protein FliF
Method: single particle / : Johnson S, Fong YH, Deme JC, Furlong EJ, Kuhlen L, Lea SM

PDB-6sd1: 
Structure of the RBM3/collar region of the Salmonella flagella MS-ring protein FliF with 33-fold symmetry applied
Method: single particle / : Johnson S, Fong YH, Deme JC, Furlong EJ, Kuhlen L, Lea SM

PDB-6sd2: 
Structure of the RBM2inner region of the Salmonella flagella MS-ring protein FliF with 21-fold symmetry applied.
Method: single particle / : Johnson S, Fong YH, Deme JC, Furlong EJ, Kuhlen L, Lea SM

PDB-6sd3: 
34mer structure of the Salmonella flagella MS-ring protein FliF
Method: single particle / : Johnson S, Fong YH, Deme JC, Furlong EJ, Kuhlen L, Lea SM

PDB-6sd4: 
Structure of the RBM3/collar region of the Salmonella flagella MS-ring protein FliF with 34-fold symmetry applied
Method: single particle / : Johnson S, Fong YH, Deme JC, Furlong EJ, Kuhlen L, Lea SM

PDB-6sd5: 
Structure of the RBM2 inner ring of Salmonella flagella MS-ring protein FliF with 22-fold symmetry applied
Method: single particle / : Johnson S, Fong YH, Deme JC, Furlong EJ, Kuhlen L, Lea SM

PDB-6tre: 
Structure of the RBM3/collar region of the Salmonella flagella MS-ring protein FliF with 32-fold symmetry applied
Method: single particle / : Johnson S, Fong YH, Deme JC, Furlong EJ, Kuhlen L, Lea SM

EMDB-6457: 
Cryo-EM of CHIKV VLP in complex with Fab fragment of neutralizing antibody IM-CKV063
Method: single particle / : Jin J, Liss N, Chen DH, Liao M, Fox JM, Shimak RM, Fong RH, Chafets D, Bakkour S, Keating S, Fomin ME, Muench MO, Sherman MB, Doranz BJ, Diamond MS, Simmons G

EMDB-6466: 
Cryo-EM of CHIKV VLP
Method: single particle / : Jin J, Liss N, Chen DH, Liao M, Fox JM, Shimak RM, Fong RH, Chafets D, Bakkour S, Keating S, Fomin ME, Muench MO, Sherman MB, Doranz BJ, Diamond MS, Simmons G

EMDB-6467: 
Cryo-EM of CHIKV VLP in complex with Fab fragment of neutralizing antibody C9
Method: single particle / : Jin J, Liss N, Chen DH, Liao M, Fox JM, Shimak RM, Fong RH, Chafets D, Bakkour S, Keating S, Fomin ME, Muench MO, Sherman MB, Doranz BJ, Diamond MS, Simmons G

EMDB-3144: 
Electron cryo-microscopy of chikungunya virus in complex with neutralizing antibody Fab CHK265
Method: single particle / : Fox JM, Long F, Edeling MA, Lin H, Duijl-Richter M, Fong RH, Kahle KM, Smit JM, Jin J, Simmons G, Doranz BJ, Crowe JE, Fremont DH, Rossmann MG, Diamond MS

PDB-5any: 
Electron cryo-microscopy of chikungunya virus in complex with neutralizing antibody Fab CHK265
Method: single particle / : Fox JM, Long F, Edeling MA, Lin H, Duijl-Richter M, Fong RH, Kahle KM, Smit JM, Jin J, Simmons G, Doranz BJ, Crowe JE, Fremont DH, Rossmann MG, Diamond MS

EMDB-3148: 
Electron cryo-microscopy of chikungunya virus in complex with neutralizing antibody Fab 4J21
Method: single particle / : Long F, Fong RH, Austin SK, Chen Z, Klose T, Fokine A, Liu Y, Porta J, Sapparapu G, Akahata W, Doranz BJ, Crowe JE, Diamond MS, Rossmann MG

EMDB-3149: 
Electron cryo-microscopy of chikungunya virus in complex with neutralizing antibody Fab 5M16
Method: single particle / : Long F, Fong RH, Austin SK, Chen Z, Klose T, Fokine A, Liu Y, Porta J, Sapparapu G, Akahata W, Doranz BJ, Crowe JE, Diamond MS, Rossmann MG

EMDB-6495: 
Electron microscopy of apo human XPC complex
Method: single particle / : Zhang ET, He Y, Grob P, Fong YW, Nogales E, Tjian R

EMDB-6496: 
Electron microscopy of apo yeast Rad4 (XPC homolog) complex
Method: single particle / : Zhang ET, He Y, Grob P, Fong YW, Nogales E, Tjian R

EMDB-6497: 
Electron microscopy of apo human XPC complex
Method: single particle / : Zhang ET, He Y, Grob P, Fong YW, Nogales E, Tjian R

EMDB-6498: 
Electron microscopy of DNA-bound human XPC complex
Method: single particle / : Zhang ET, He Y, Grob P, Fong YW, Nogales E, Tjian R

EMDB-2799: 
Cryo-EM structure of gamma-TuSC oligomers in a closed conformation
Method: helical / : Kollman JM, Greenberg CH, Li S, Moritz M, Zelter A, Fong K, Fernandez JJ, Sali A, Kilmartin J, Davis TN, Agard DA

EMDB-5989: 
Electron cryo-tomography of the Microtubule Nucleation Complex in the Yeast Spindle Pole Body
Method: subtomogram averaging / : Kollman JM, Greenberg C, Li S, Fernandez JJ, Moritz M, Zelter A, Fong K, Sali A, Kilmartin JV, Davis TN, Agard DA
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
