-Search query
-Search result
Showing 1 - 50 of 373 items for (author: briggs & j)

EMDB-19024: 
Structure of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG
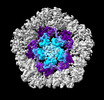
EMDB-19025: 
Structure of the five-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG

EMDB-19026: 
Structure of the three-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG

EMDB-19027: 
Structure of the two-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG

PDB-8rb3: 
Structure of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG

PDB-8rb4: 
Structure of the five-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG

PDB-8rb5: 
Structure of the three-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG

PDB-8rb7: 
Structure of the two-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG
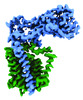
EMDB-41299: 
Structural basis of peptidoglycan synthesis by E. coli RodA-PBP2 complex
Method: single particle / : Nygaard R, Mancia F

EMDB-41303: 
Transmembrane map
Method: single particle / : Nygaard R, Mancia F
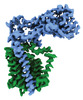
EMDB-41304: 
Periplasmic map
Method: single particle / : Nygaard R, Mancia F

PDB-8tj3: 
Structural basis of peptidoglycan synthesis by E. coli RodA-PBP2 complex
Method: single particle / : Nygaard R, Mancia F

EMDB-16698: 
Subtomogram average of HIV-1 CA pentamer from capsid-like particles assembled with inositol hexakisphosphate
Method: subtomogram averaging / : Tan A, Briggs JAG, Dick RA

EMDB-16699: 
Subtomogram average of HIV-1 CA hexamer from capsid-like particles assembled with inositol hexakisphosphate
Method: subtomogram averaging / : Tan A, Briggs JAG, Dick RA

EMDB-29772: 
HIV-1 CA lattice bound to IP6; from capsid-like particles
Method: single particle / : Highland CM, Dick RA

EMDB-29773: 
HIV-1 capsid lattice bound to IP6, pH 6.2
Method: single particle / : Highland CM, Dick RA

EMDB-29774: 
HIV-1 CA lattice bound to IP6, pH 7.4
Method: single particle / : Highland CM, Dick RA

EMDB-29775: 
HIV-1 capsid lattice bound to dNTPs
Method: single particle / : Highland CM, Dick RA

EMDB-29776: 
HIV-1 capsid lattice bound to IP6 and Lenacapavir
Method: single particle / : Highland CM, Dick RA

EMDB-29777: 
Defective HIV-1 CA pentamer in surrounding hexameric lattice
Method: single particle / : Highland CM, Dick RA

PDB-8g6k: 
HIV-1 CA lattice bound to IP6; from capsid-like particles
Method: single particle / : Highland CM, Dick RA

PDB-8g6l: 
HIV-1 capsid lattice bound to IP6, pH 6.2
Method: single particle / : Highland CM, Dick RA

PDB-8g6m: 
HIV-1 CA lattice bound to IP6, pH 7.4
Method: single particle / : Highland CM, Dick RA

PDB-8g6n: 
HIV-1 capsid lattice bound to dNTPs
Method: single particle / : Highland CM, Dick RA

PDB-8g6o: 
HIV-1 capsid lattice bound to IP6 and Lenacapavir
Method: single particle / : Highland CM, Dick RA

EMDB-16703: 
HIV-1 mature capsid hexamer from CA-IP6 CLPs
Method: single particle / : Stacey JCV, Briggs JAG

EMDB-16704: 
HIV-1 mature capsid pentamer from CA-IP6 CLPs
Method: single particle / : Stacey JCV, Briggs JAG

EMDB-16705: 
HIV-1 mature capsid hexamer next to pentamer (type I) from CA-IP6 CLPs
Method: single particle / : Stacey JCV, Briggs JAG

EMDB-16706: 
HIV-1 mature capsid hexamer from CA-IP6 CLPs, bound to Nup153 peptide
Method: single particle / : Stacey JCV, Briggs JAG

EMDB-16707: 
HIV-1 mature capsid pentamer from CA-IP6 CLPs bound to Nup153 peptide
Method: single particle / : Stacey JCV, Briggs JAG

EMDB-16708: 
HIV-1 mature capsid hexamer next to pentamer (type I) from CA-IP6 CLPs bound to Nup153 peptide.
Method: single particle / : Stacey JCV, Briggs JAG

EMDB-16709: 
HIV-1 mature capsid hexamer from CA-IP6 CLPs, bound to CPSF6 peptide.
Method: single particle / : Stacey JCV, Briggs JAG

EMDB-16710: 
HIV-1 mature capsid pentamer from CA-IP6 CLPs bound to CPSF6 peptide
Method: single particle / : Stacey JCV, Briggs JAG

EMDB-16711: 
HIV-1 mature capsid hexamer from CA-IP6 CLPs, bound to Sec24C peptide.
Method: single particle / : Stacey JCV, Briggs JAG

EMDB-16712: 
HIV-1 mature capsid pentamer from CA-IP6 CLPs bound to Sec24C peptide
Method: single particle / : Stacey JCV, Briggs JAG

PDB-8ckv: 
HIV-1 mature capsid hexamer from CA-IP6 CLPs
Method: single particle / : Stacey JCV, Briggs JAG

PDB-8ckw: 
HIV-1 mature capsid pentamer from CA-IP6 CLPs
Method: single particle / : Stacey JCV, Briggs JAG

PDB-8ckx: 
HIV-1 mature capsid hexamer next to pentamer (type I) from CA-IP6 CLPs
Method: single particle / : Stacey JCV, Briggs JAG

PDB-8cky: 
HIV-1 mature capsid hexamer from CA-IP6 CLPs, bound to Nup153 peptide
Method: single particle / : Stacey JCV, Briggs JAG

PDB-8ckz: 
HIV-1 mature capsid pentamer from CA-IP6 CLPs bound to Nup153 peptide
Method: single particle / : Stacey JCV, Briggs JAG

PDB-8cl0: 
HIV-1 mature capsid hexamer next to pentamer (type I) from CA-IP6 CLPs bound to Nup153 peptide.
Method: single particle / : Stacey JCV, Briggs JAG

PDB-8cl1: 
HIV-1 mature capsid hexamer from CA-IP6 CLPs, bound to CPSF6 peptide.
Method: single particle / : Stacey JCV, Briggs JAG

PDB-8cl2: 
HIV-1 mature capsid pentamer from CA-IP6 CLPs bound to CPSF6 peptide
Method: single particle / : Stacey JCV, Briggs JAG

PDB-8cl3: 
HIV-1 mature capsid hexamer from CA-IP6 CLPs, bound to Sec24C peptide.
Method: single particle / : Stacey JCV, Briggs JAG

PDB-8cl4: 
HIV-1 mature capsid pentamer from CA-IP6 CLPs bound to Sec24C peptide
Method: single particle / : Stacey JCV, Briggs JAG

EMDB-16183: 
In situ structure of the Caulobacter crescentus S-layer
Method: subtomogram averaging / : von Kuegelgen A, Bharat T

EMDB-16207: 
HIV-1 CA-SP1 subtomogram average with Relion4 from EMPIAR-10164 dataset, 3.2 A from 5 tomograms
Method: subtomogram averaging / : Ke Z, Briggs JAG, Scheres SHW

EMDB-16209: 
HIV-1 CA-SP1 subtomogram average with Relion4 from EMPIAR-10164 dataset, 3.0 A from the full dataset
Method: subtomogram averaging / : Ke Z, Briggs JAG, Scheres SHW

PDB-8bqe: 
In situ structure of the Caulobacter crescentus S-layer
Method: subtomogram averaging / : von Kuegelgen A, Bharat T
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
